Application of saccharomyces rouxii gene in improving yield of HDMF produced by microorganisms
A technology of Saccharomyces ruckeri and microorganisms, applied in the field of microorganisms, can solve the problems of unsuitable factory production and high price, and achieve the effects of high safety, good fragrance and environmental friendliness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
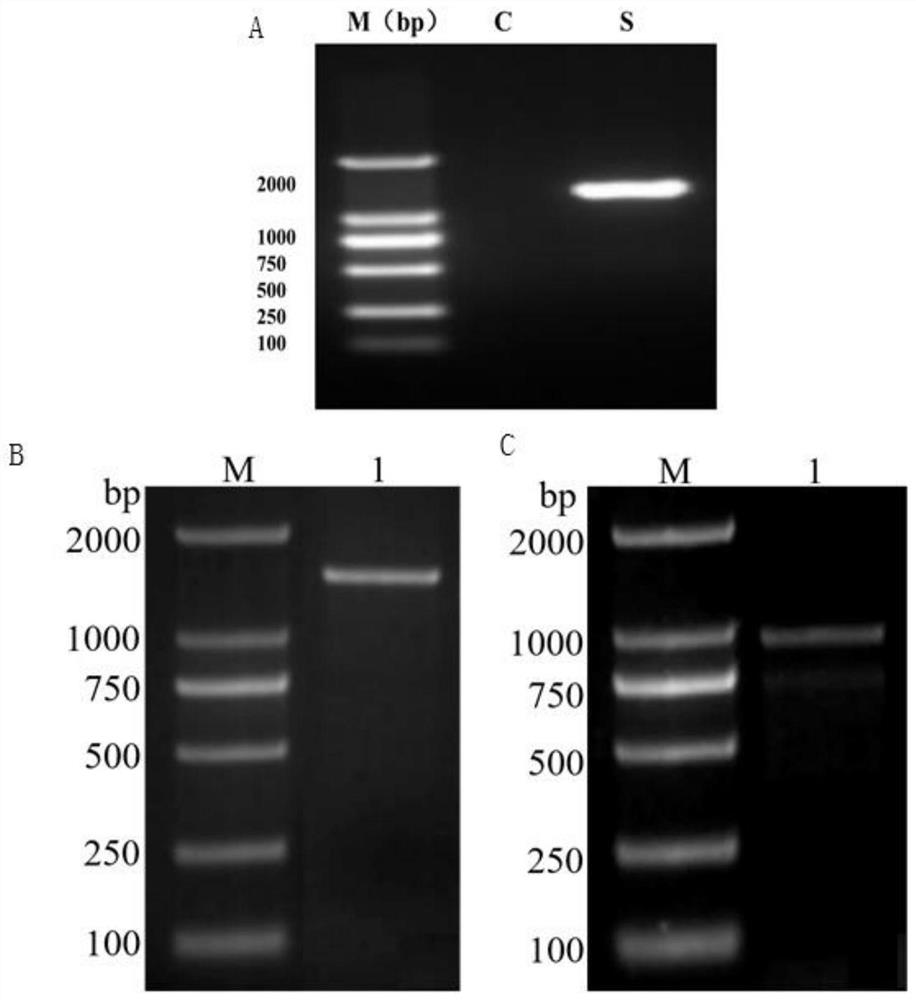
[0035] 1.1 Construction of QOR gene overexpression recombinant plasmid, FBA gene and TPI gene overexpression recombinant plasmid
[0036] Using the Saccharomyces ruxii genome as a template, gene amplification primers are shown in Table 1, and KOD FX Neo high-fidelity enzyme was used to perform QOR gene ( figure 1 A), FBA gene ( figure 1 B) and the cloning of TPI gene ( figure 1C), the successfully cloned QOR gene, FBA gene and TPI gene target fragments were sent to Shanghai Sangon Biotechnology Co., Ltd. for sequencing, and DNAMAN 7.0 was used to splice the sequencing results. The obtained sequence was aligned with the QOR (XM_002494472.1), FBA (XM_002499162.1) and TPI (XM_002498482.1) sequences in the NCBI genome of Luxie yeast.
[0037] The sequence of the QOR gene coding region is as follows:
[0038] ATGCTTAGATCGACAATATCCTCAGTGTTCAACAAGAGGGCAACAATGGCAACCACAACTATTCCAAAGACCCAAAAAGTTATTTTGATTAACGATACTGGTAGTTATGATGTGCTGAAATACCAGGATCATCCGGTGCCTAGTATTACTGACAATGAATTATTGGTTAAAA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com