Isolated polypeptide, nucleic acid and application thereof
A nucleic acid and nucleotide sequence technology, applied in the field of biochemistry, can solve the problems of non-conformity with green chemistry, high energy consumption, harsh reaction conditions, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0104] 1. Construction of recombinant plasmids
[0105] 1. For the E. coli system, optimize the codons of the β-aminopeptidase sequence gene found in the deep-sea metagenomic library, and synthesize the related gene (SEQ ID NO: 4).
[0106] 2. Take the gene synthesized in step 1, perform double digestion with restriction endonucleases NcoI and XhoI, and recover the digested product.
[0107] 3. Take the pET-26b plasmid, perform double digestion with restriction endonucleases NcoI and XhoI, and recover the vector skeleton.
[0108] 4. Ligate the digested product of step 2 with the vector backbone of step 3 to obtain the recombinant plasmid pET26b-WTCar. According to the sequencing results, the structure of the recombinant plasmid pET26b-WTCar is described as follows: a DNA molecule shown in SEQ ID NO:4 is inserted between the NcoI and XhoI restriction sites of the pET-26b plasmid. The DNA molecule shown in SEQ ID NO:4 encodes the wild-type protein shown in SEQ ID NO:1.
[01...
Embodiment 2
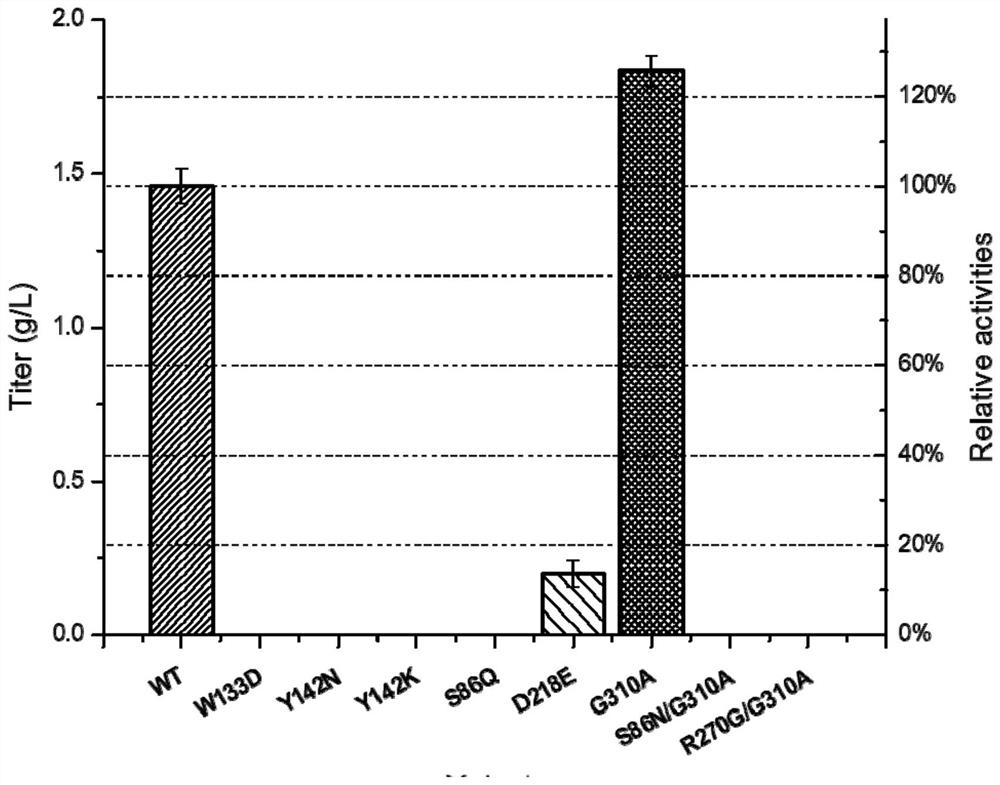
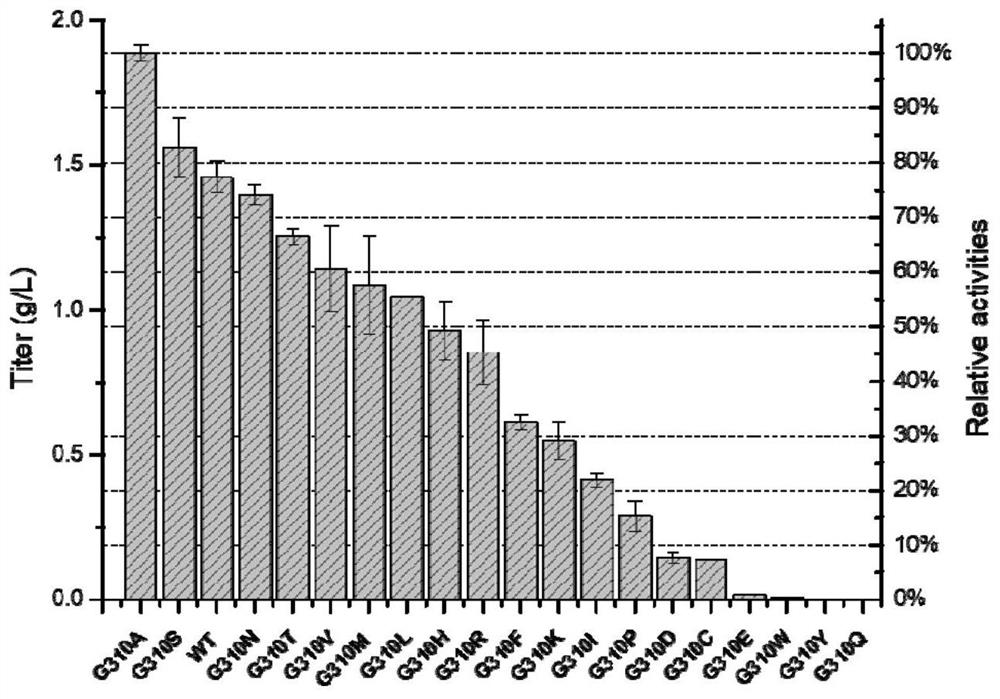
[0163] Example 2. Whole-cell catalysis and enzyme activity detection of wild-type protein and each mutant protein
[0164] 1. Protein expression
[0165] Each of the recombinant bacteria prepared in Example 1 was used to prepare each protein with a His6 tag fused at the N-terminal, and the specific steps were as follows
[0166] 1. Inoculate the recombinant bacteria into a liquid LB medium containing 40 μg / mL kanamycin, and culture it with shaking at 37° C. and 200 rpm until the OD600nm value = 0.6-0.8.
[0167] 2. After completing step 1, add isopropyl-β-D-thiogalactopyranoside (IPTG) to the system at a concentration of 0.02mM, shake and culture at 16°C and 200rpm for 8 hours.
[0168] 3. After completing step 2, collect the cells by centrifugation.
[0169] 4. Take the cells obtained in step 3, suspend them with binding buffer, then perform ultrasonic disruption (10% power, break for 3 seconds and stop for 5 seconds, total time 10 minutes), then centrifuge at 12000g for 20...
Embodiment 3
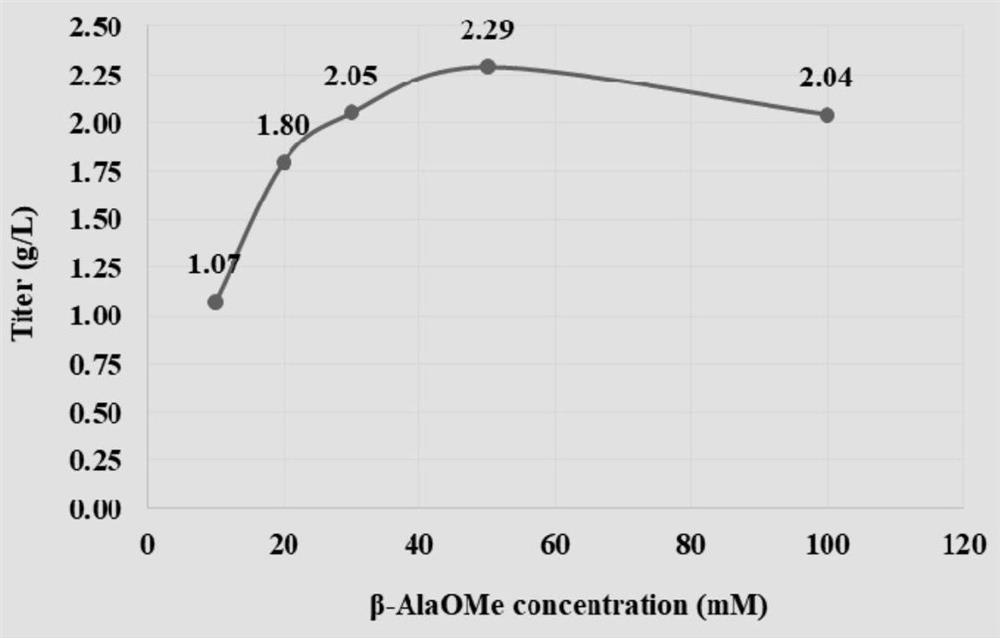
[0187] Embodiment 3, the impact of G310A mutant whole cell catalytic synthesis of L-carnosine
[0188] In order to further improve the enzyme activity of G310A and simplify the process flow, L-carnosine was catalyzed and synthesized by whole cells of G310A mutant, and the reaction conditions were optimized.
[0189] 1. Preparation of whole cell extracts
[0190] The G310A prepared in Example 1 was used to prepare the whole cell extract, and the specific steps were as follows:
[0191] 1. Inoculate the recombinant bacteria into a liquid LB medium containing 40 μg / mL kanamycin, and culture it with shaking at 37° C. and 200 rpm until the OD600nm value = 0.6-0.8.
[0192]2. After completing step 1, add isopropyl-β-D-thiogalactopyranoside (IPTG) to the system at a concentration of 0.02mM, shake and culture at 16°C and 200rpm for 8 hours.
[0193] 3. After completing step 2, collect the cells by centrifugation.
[0194] 4. Take the cells obtained in step 3, wash twice with Na2CO3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com