Device and method for high-purity sorting of rare cells
A rare cell, high-purity technology, applied in the field of devices for high-purity sorting of rare cells, can solve the problems of inability to apply cell sorting, low sorting speed and efficiency, complex structure and operation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0084] As a specific implementation, the rare cell sorting method is as follows:
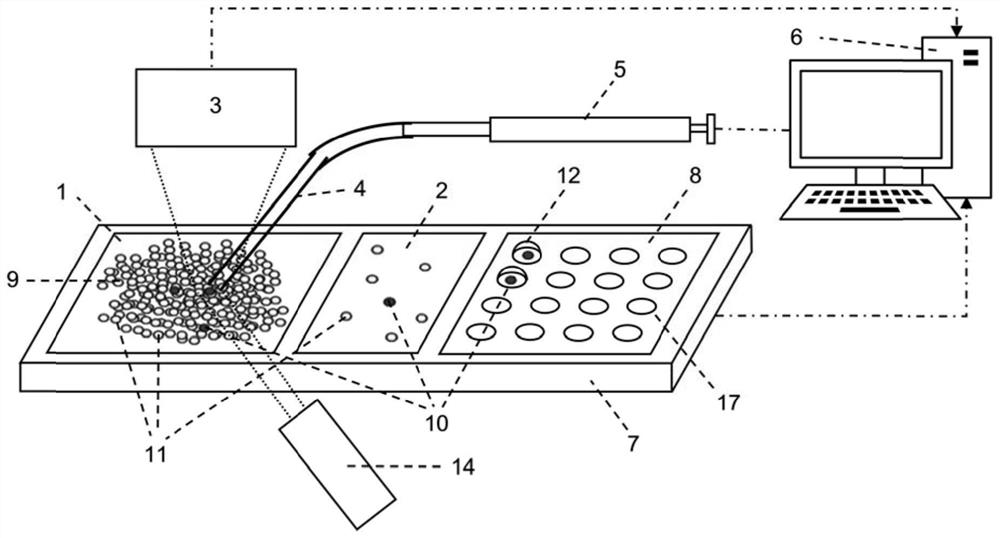
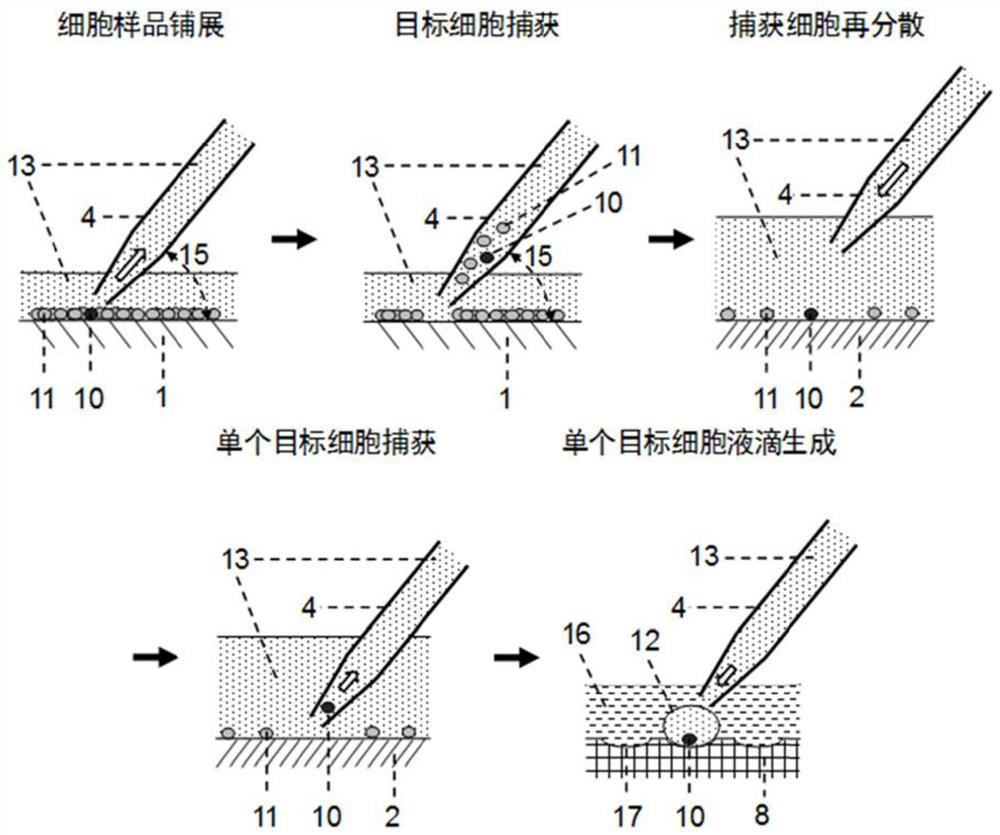
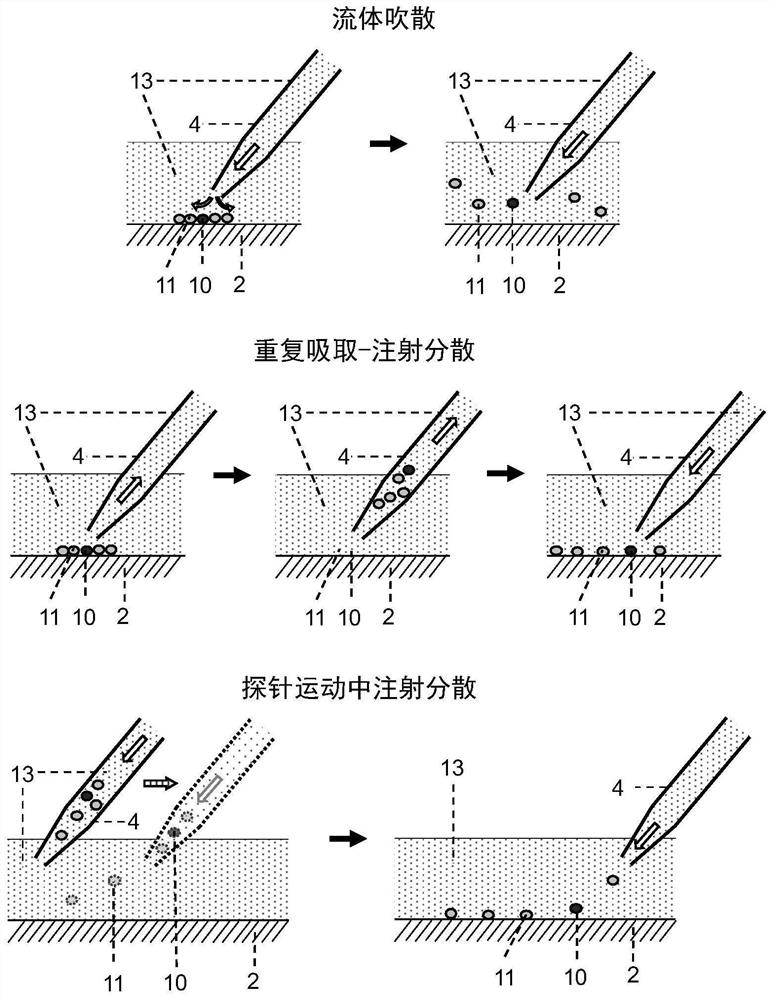
[0085] Clean flat-bottomed glass containers with ethanol, water, and EDTA in sequence (you can use figure 1 The integrated flat-bottomed container shown can also be a separately set flat-bottomed container, such as a petri dish, etc.), and the cell suspension (that is, the suspension of cell sample 9) that has been labeled with a fluorescent antibody into the flat-bottomed glass container In step 1, oscillate to disperse the cells evenly, settle naturally for 3-5 minutes, and spread the cells into a cell layer within 3 layers ( figure 2 ).
[0086] Fluorescently label target cells with specific antibodies before detection, such as Figure 4 As shown, using Cell Imaging Detector 3 (Laser Induced Fluorescence Cell Imaging Detector, such as Figure 4 ) with the use of light source 14 to scan the fluorescent signal in the detection area, and use the computer vision software based on threshold a...
Embodiment 2
[0092] In embodiment 2, device structure is as Figure 7 shown.
[0093] Pour the cell suspension containing the target cells 10 labeled with fluorescent antibodies and non-target interfering cells 11 into the culture dish 1, and incubate in the incubator for about 30 minutes. At this time, the adherent cells can form a tightly packed monolayer Adherent growth. Add an appropriate amount of trypsin cell digestion solution to digest the cells and form a low-adhesion single-cell layer at the bottom of the culture dish.
[0094] Use the motorized microscope 3 to scan the bright field and fluorescence images of the detection area, and use the image classification software based on artificial intelligence to automatically identify the fluorescent cells therein (see Example 1). Synthesize the fluorescence image recognition results under different excitation wavelengths to determine the identity of the target cells location and count its number. It is also possible to combine multi...
Embodiment 3
[0100] In embodiment 3, device structure is as Figure 9 shown.
[0101] The integrated chip 19 is used for experiments, and the integrated chip is processed with a cell spreading area (equivalent to container 1) and a multiple dispersion area (equivalent to container 2) with microstructures (including micro-pit arrays and micro-pillar arrays 18) arrays. and the area carrying the captured cell droplet (equivalent to chip 8). Pour the cell suspension containing the target cells into the cell spreading area of the integrated chip, and through natural sedimentation and scraper control, a monolayer of cell spreading is formed, and the cells are bound in the micropit or the gap between the micropillars. The surface of the sample scanning area is modified in advance to inhibit the non-specific adsorption of proteins and reduce the adhesion of cells on its surface.
[0102] Use the fast and high-sensitivity cell imaging detector 3 (laser-induced fluorescence cell imaging detector...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com