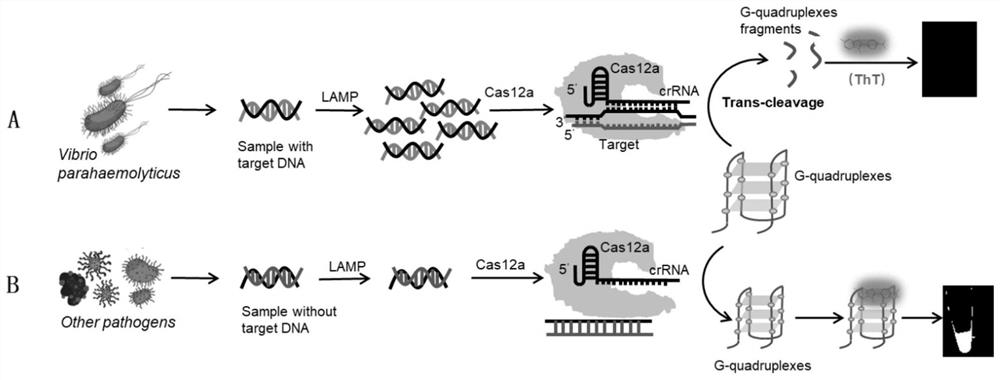
Method for label-free visual detection of vibrio parahaemolyticus genes based on CRISPR(clustered regularly interspaced short palindromic repeats)/Cas12a
A hemolytic vibrio, label-free technology, applied in the field of microbial detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
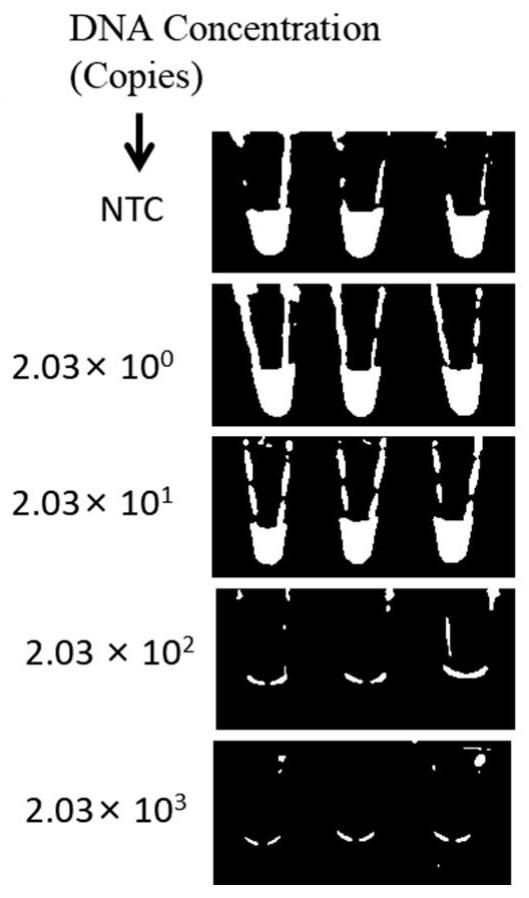
[0062] Embodiment 1: sensitivity test
[0063] Genomic DNA of Vibrio parahaemolyticus ATCC 17802 was extracted using a commercially available bacterial genomic DNA extraction kit (Beijing Biotech Biotechnology Co., Ltd.), its concentration was measured by a micro-volume ultraviolet spectrophotometer (Thermo Fisher, USA), and the copy number was calculated.
[0064] The extracted Vibrio parahaemolyticus genomic DNA was diluted tenfold with sterile water so that its concentration was 10 4 , 10 3 , 10 2 , 10 1 Copies / μL (copies / μL).
[0065] Using the gradient diluted Vibrio parahaemolyticus genomic DNA as a template, use LAMP to amplify: 12.5 μL amplification system includes 2 μL of Vibrio parahaemolyticus DNA template, 1.6 μM internal primer 1 and internal primer 2, and 0.2 μM external primer 1 and outer primer 2, 0.4μM loop primer 1 and loop primer 2, 1.4mM dNTPs, 8U Bst DNA Polymerase Large Fragment, 1×NEBuffer2.1, and incubated at 65°C for 60min to obtain the LAMP produc...
Embodiment 2
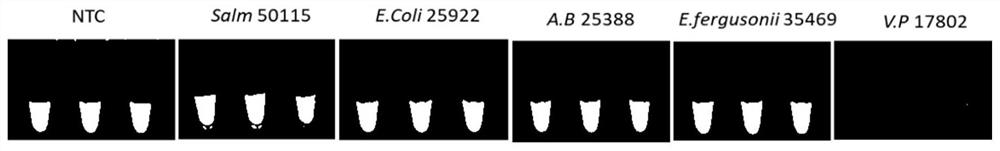
[0067] Embodiment 2: specificity experiment
[0068]Using a commercially available bacterial genomic DNA extraction kit (Beijing Baitaike Biotechnology Co., Ltd.) from Vibrio parahaemolyticus ATCC 17802, Salmonella typhimurium CMCC (B) 50115 (Salmonella 50115), Acinetobacter DSM 25388 (Acinetobacter 25388), Genomic DNA was extracted from Escherichia ferguson 056 (Escherichia ferguson 056) and Escherichia coli ATCC25922 (Escherichia coli 25922), and its concentration was measured by a micro-ultraviolet spectrophotometer (ThermoFisher, USA) to calculate the copy number.
[0069] Using the DNA of the above five kinds of bacteria as templates, amplified with Tlh primers respectively to obtain five kinds of amplification products: 12.5 μL amplification system including 2 μL DNA templates corresponding to bacteria, 1.6 μM inner primer 1 and inner primer 2, 0.2 μM outer primer Primer 1 and outer primer 2, 0.4 μM loop primer 1 and loop primer 2, 1.4 mM dNTPs, 8U Bst DNA Polymerase Lar...
Embodiment 3
[0072] Example 3: Detection of Vibrio parahaemolyticus in fresh fresh shrimp
[0073] The preserved strain of Vibrio parahaemolyticus was separated by streaking on the plate medium, and then a single colony was picked and transferred to LB broth liquid medium, cultured at 37°C for 6 hours, diluted 10 times with normal saline, and calculated according to the plate count Contains 6.1×10 5 CFU / mL to 6.1×10 0 CFU / mL of Vibrio parahaemolyticus. Dip the aseptic prawns into the diluent to infect the bacteria, and then let them stand at room temperature for 30 minutes in a sterile environment to make Vibrio parahaemolyticus tightly adhere to the surface of the prawns.
[0074] The prawns contaminated by the above-mentioned Vibrio parahaemolyticus were used as samples, and DNA was extracted from them using a commercially available bacterial genomic DNA extraction kit (Beijing Biotech Biotechnology Co., Ltd.).
[0075] The DNA extracted from the polluted prawns was used as a template...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com