Preparation method for circular single-stranded DNA integrated with aptamer and applications of circular single-stranded DNA integrated with aptamer in DNA origami
A nucleic acid aptamer and aptamer technology, applied in the fields of DNA nanotechnology and biological applications, can solve the problems of low ligation efficiency and stability, and achieve the effects of efficient preparation and rich diversity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
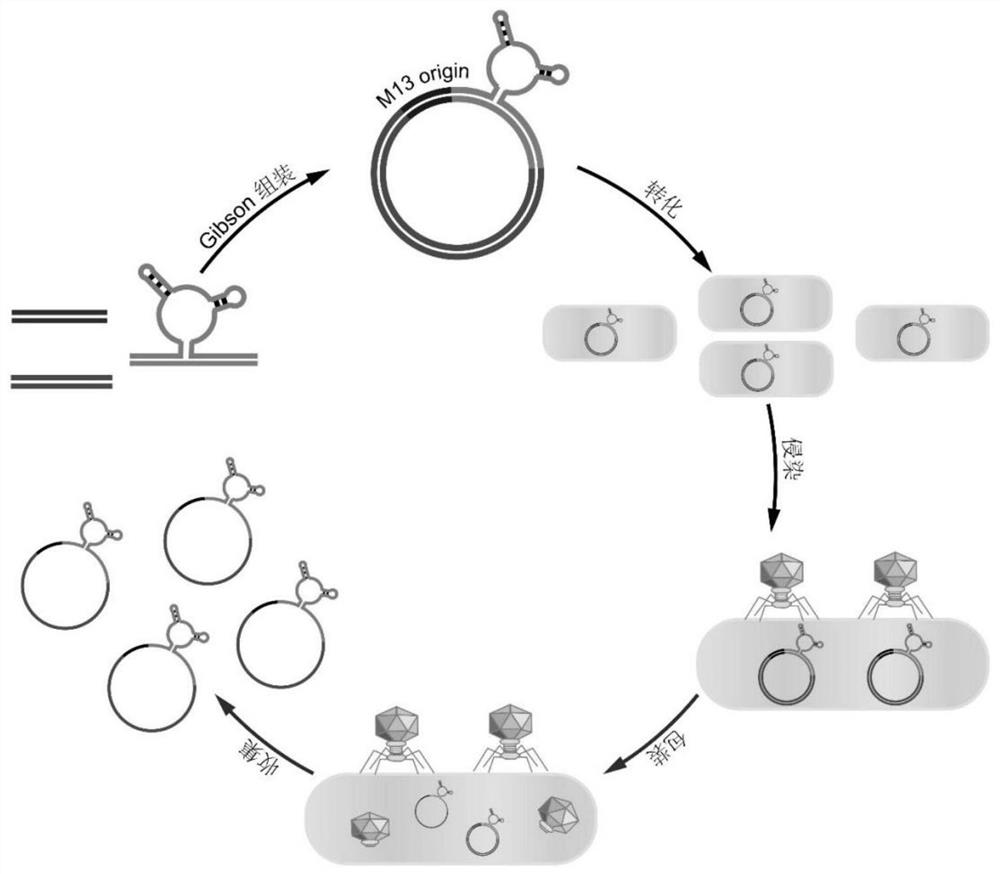
[0028] As shown in the figure; a method for preparing a circular single-stranded DNA integrated with a nucleic acid aptamer, and using the obtained circular single-stranded DNA integrated with a nucleic acid aptamer to construct a functional DNA origami structure, the specific steps are as follows:
[0029] (1.1), constructing circular double-stranded recombinant phagemids containing nucleic acid aptamer sequences;
[0030] (1.2), transforming the constructed circular double-stranded recombinant phagemid into Escherichia coli competent cells; extracting circular single-stranded DNA with nucleic acid aptamer integration.
[0031] Further, in the step (1.1), the specific operation method for constructing a circular double-stranded recombinant phagemid containing a nucleic acid aptamer sequence is as follows:
[0032] First, the selected nucleic acid adapter fragments are obtained by using chemical synthesis, and the nucleic acid adapter fragments are connected to the DNA fragmen...
Embodiment 1
[0043] Preparation of ssDNA integrated with human α-thrombin aptamer TBA-15 and TBA-29 and PDGF aptamer;
[0044] Utilizing the present invention, the preparation of human α-thrombin aptamer TBA-15 and ssDNA integrating TBA-29 and PDGF aptamer comprises the following steps:
[0045] (1), constructing a circular double-stranded recombinant phagemid containing a nucleic acid aptamer sequence;
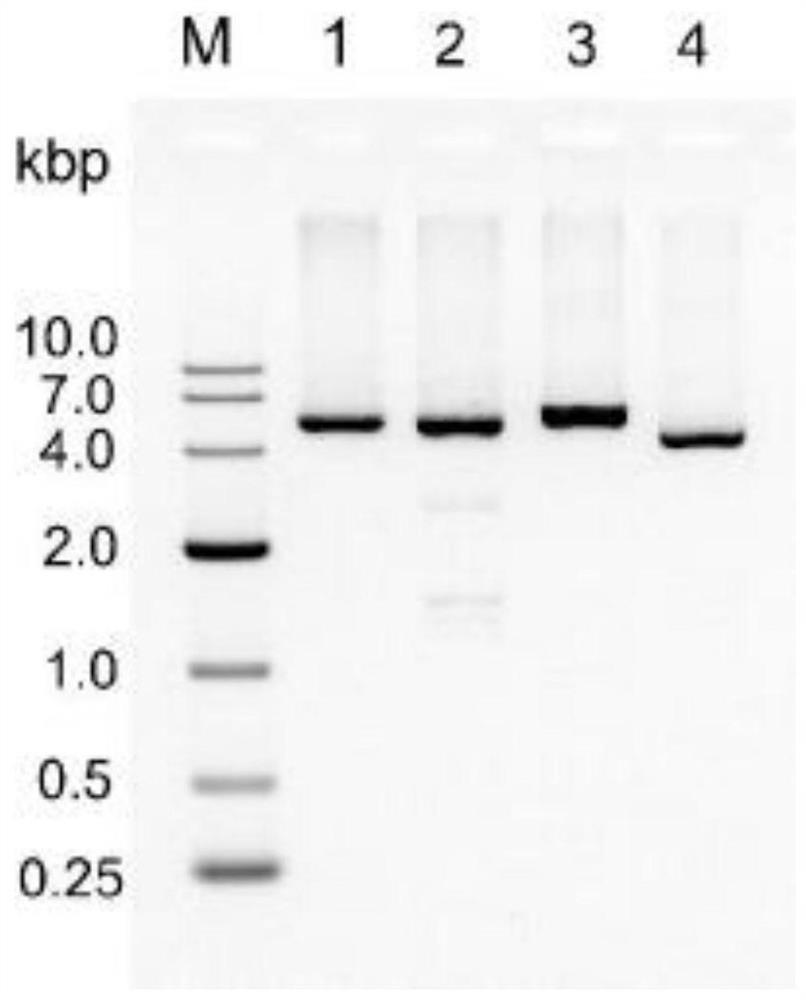
[0046] Human α-thrombin aptamers TBA-15 and TBA-29 and PDGF aptamer fragments were obtained by chemical synthesis; two human α-thrombin aptamers TBA-15 and TBA-29 were synthesized by overlapping PCR. The PDGF aptamer fragment (sequence shown in Table 1) was connected to the DNA fragment of the selected region in the M13mp18 RF DNA vector; the product of overlapping PCR was purified by agarose gel electrophoresis and then assembled by Gibson The method was inserted into the M13mp18 RF DNA vector and replaced the original DNA fragment in this region, and verified by gene sequencing, the re...
Embodiment 2
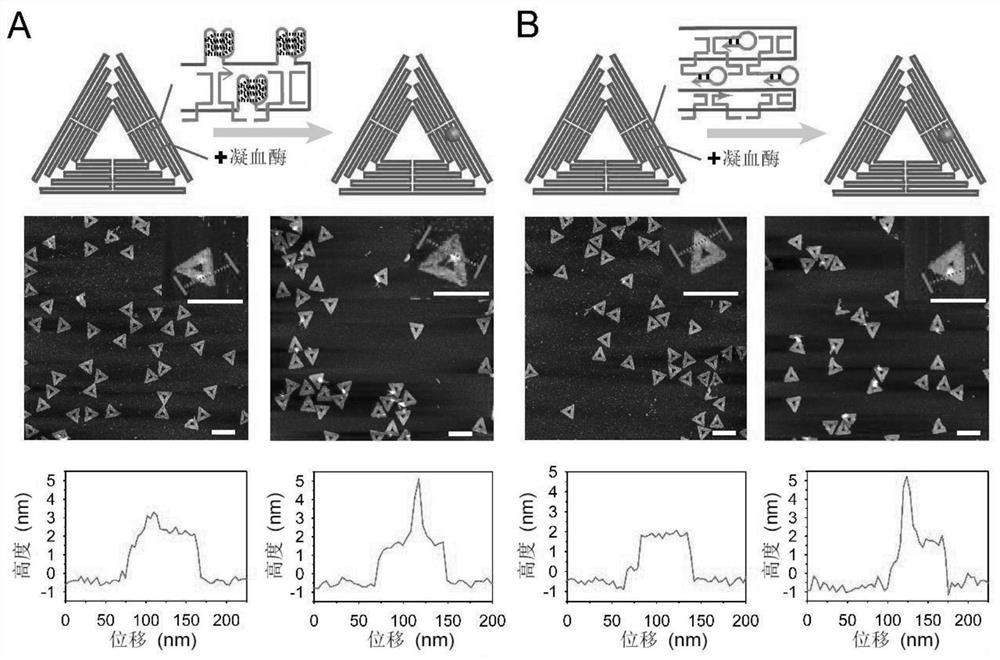
[0052] Example 2. Constructing a functional DNA origami structure of nucleic acid aptamers integrating backbone chains:
[0053] Using the ssDNA integrated with the nucleic acid aptamer prepared by the present invention to construct a functional DNA origami structure, the specific operations are as follows:
[0054] Mix the circular ssDNA integrated with different aptamers prepared in Example 1 with their corresponding staple strands at a molar ratio of 1:10; according to the requirements of subsequent experiments, the final concentration of ssDNA is in the range of 1-10 nM; add A certain amount of 1×TAE / Mg 2+ Buffer (Mg 2+ The concentration is 12.5mM) to 100μL and mixed well, then placed in the PCR instrument, set the program gradient annealing from 85°C to 25°C for 16h; Purified by agarose gel electrophoresis.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com