Method for jointly detecting whole genome DNA adduct based on dot blot hybridization and chromatin co-immunoprecipitation sequencing
A technology of co-immunoprecipitation and dot hybridization, which is applied in the field of identification and detection of BPDE adduct genes, can solve the problems of loss of DNA adducts, loss of information, failure to reflect the formation of BPDE-DNA adducts, etc., and achieve rich information Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
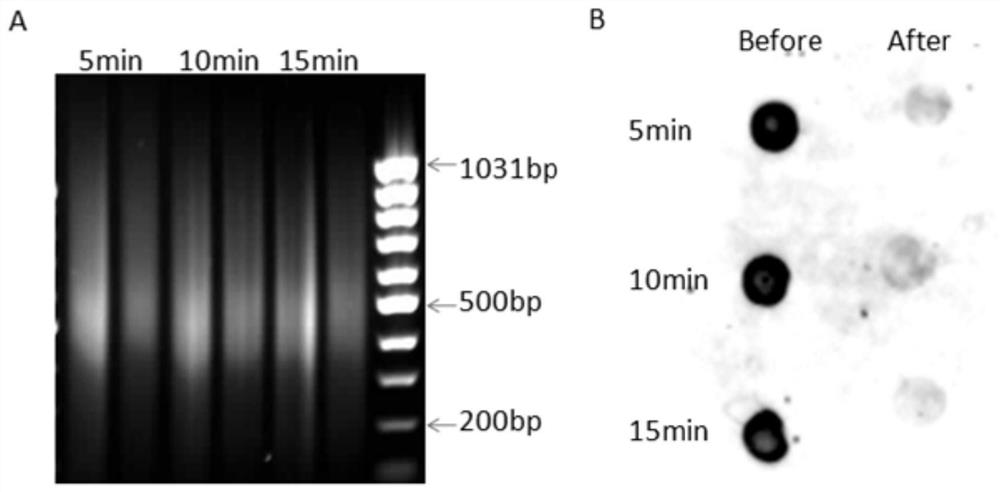
[0062] A method for joint detection of whole-genome DNA adducts based on dot blot hybridization and chromatin immunoprecipitation sequencing, comprising the following steps:
[0063] (1) Experimental method
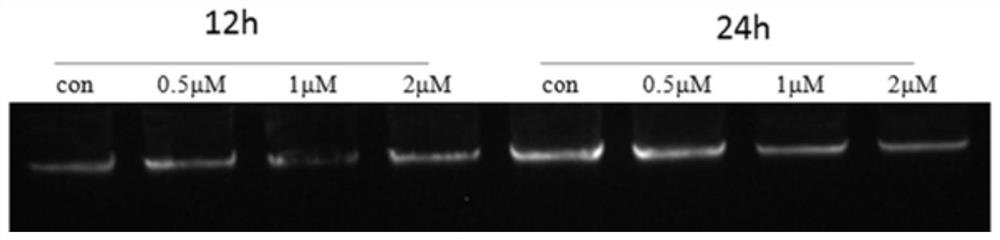
[0064]1. Design of BPDE exposure
[0065] The exposure concentration was set to 0, 0.5, 1, 2, 4, 8μM, and the exposure time was set to 12h, 24h and 36h. The 1 mM BPDE stock solution was prepared into the corresponding infection concentration with DMEM / F12 complete medium containing 5% FBS. When the BEAS-2B cells were in the logarithmic growth phase and the growth confluence reached 70%, the old culture medium was discarded, washed with PBS, and the culture medium containing the corresponding concentration of BPDE was added to continue culturing the cells. The volume fraction of DMSO in the final exposure culture solution should not exceed 5 / 1000.
[0066] 2. CCK-8 detection of cell viability
[0067] Take BEAS-2B cells in the logarithmic growth phase and inoculate the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com