A PCR-independent short-fragment cloning method
A cloning method and short fragment technology, applied in the field of bioengineering, can solve the problems of needing templates, long time-consuming, complicated and time-consuming operations, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
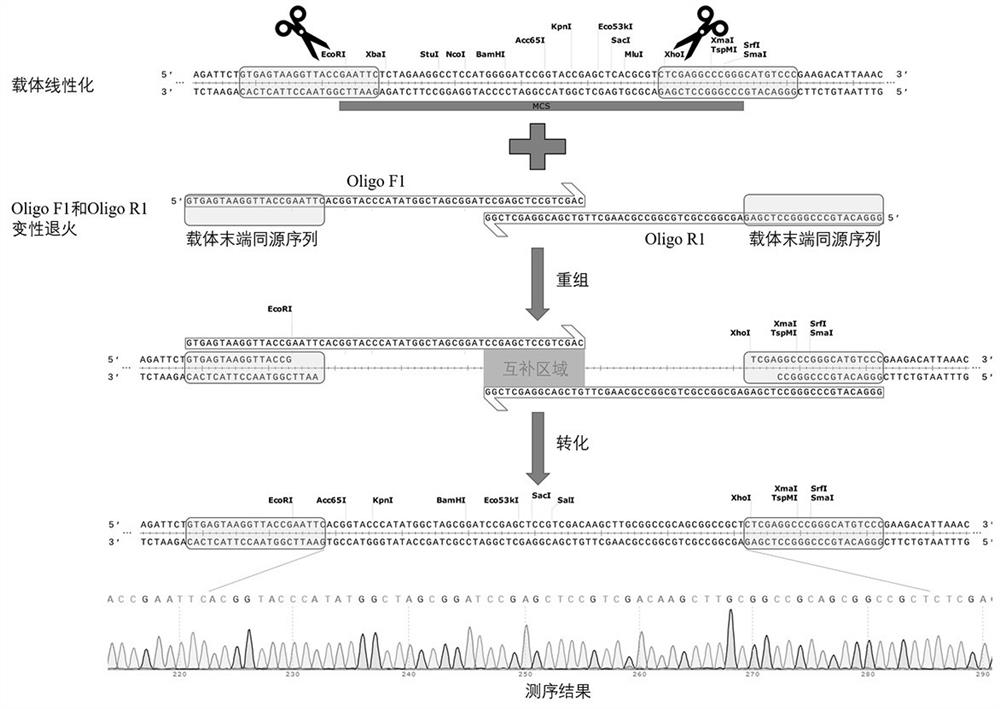
[0069] Take the restriction endonuclease site adjustment in the multiple cloning site in the gene silencing vector pTRV2 for virus induction as an example (for the specific operation process, please refer to figure 1 ), in this example, the SalI restriction site was introduced, and the arrangement order of KpnI and BamHI in the multiple cloning site was adjusted. The specific experimental process is introduced as follows.
[0070] (1) Preparation of linearized vector
[0071] The pTRV2 vector was linearized with EcoRI and XhoI, and purified and recovered. The specific operation reference is as follows:
[0072] The 50 μL digestion system was designed as follows:
[0073] pTRV2 vector, 2000 ng;
[0074] EcoRI, 2 μL;
[0075] XhoI, 2 μL;
[0076] 10x Cut smart buffer, 5 μL;
[0077] ddH 2 O, make up to 50 μL.
[0078] Digestion at 37°C overnight (about 10h), followed by purification and recovery of the digested linear vector product for later use.
[0079] (2) Design ...
Embodiment 2
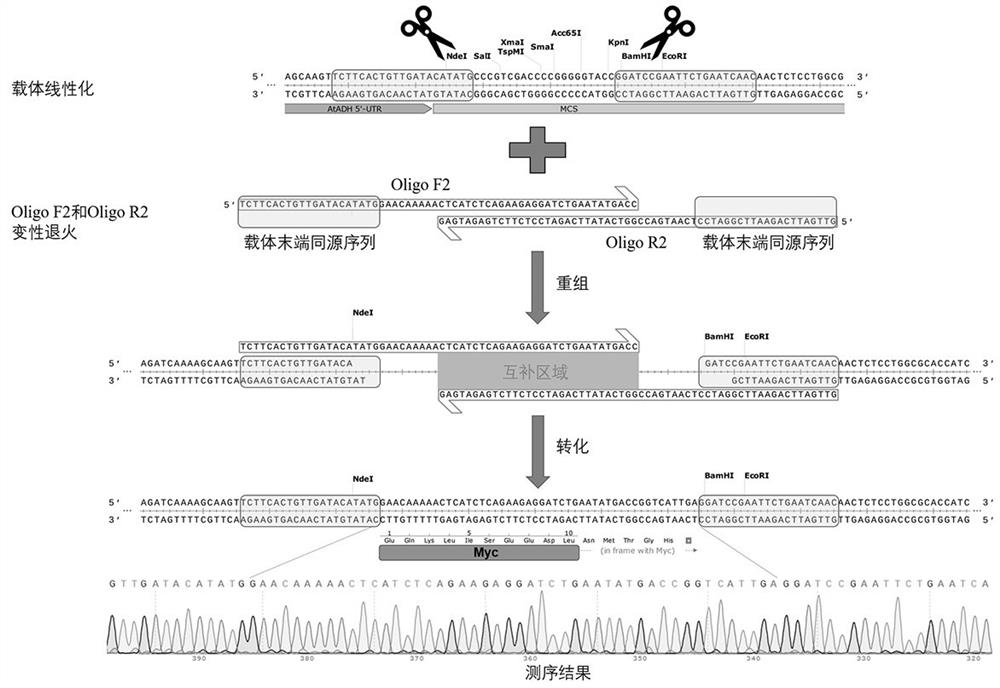
[0105] Taking the binary expression vector pRI101-AN as an example, in this example, a Myc protein tag was added to its multi-cloning site region, so that the C-terminal of the protein expressed by the subsequent transgenic plants had this tag, which was beneficial to the subsequent protein expression. Level detection (for specific operation procedures, please refer to figure 2 ).
[0106] The specific experimental process is introduced as follows.
[0107] (1) Preparation of linearized vector
[0108] The pRI101-AN vector was linearized with NdeI and BamHI, and purified and recovered. The specific operation reference is as follows:
[0109] The 50 μL digestion system is as follows:
[0110] pRI101-AN vector, 2000 ng;
[0111] NdeI, 2 μL;
[0112] BamHI, 2 μL;
[0113] 10x Cut smart buffer, 5 μL;
[0114] ddH 2 O, make up to 50 μL
[0115] Digestion at 37°C overnight (about 10h), followed by purification and recovery of the digested linear vector product for later u...
Embodiment 3
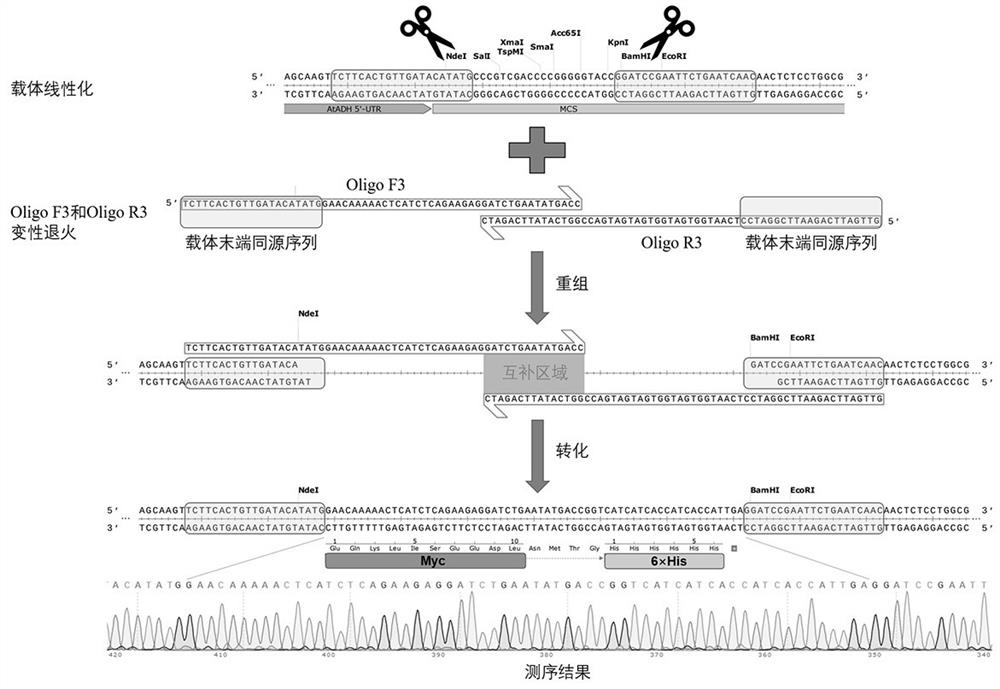
[0140] Taking the binary expression vector pRI101-AN as an example, in this example, two protein tags, Myc and His, were added to its multi-cloning site region, so that the protein C-terminus expressed by the subsequent transgenic plants had this tag, which was beneficial to the Subsequent detection of protein expression levels (for specific operation procedures, please refer to image 3 ).
[0141] (1) Preparation of linearized vector
[0142] The pRI101-AN vector was linearized with NdeI and BamHI, and purified and recovered. The specific operation reference is as follows:
[0143] The 50 μL digestion system is as follows:
[0144] pRI101-AN vector, 2000 ng;
[0145] NdeI, 2 μL;
[0146] BamHI, 2 μL;
[0147] 10x Cut smart buffer, 5 μL;
[0148] ddH 2 O, make up to 50 μL;
[0149] Digestion at 37°C overnight (about 10h), followed by purification and recovery of the digested linear vector product for later use.
[0150] (2) Design and synthesis of two long-chain oli...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com