Reagent for detecting DNA methylation and application
A methylation and gene technology, applied in recombinant DNA technology, DNA/RNA fragments, determination/inspection of microorganisms, etc., can solve the problems of decreased expression of tumor suppressor genes, decreased methylation degree, and altered chromatin structure.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
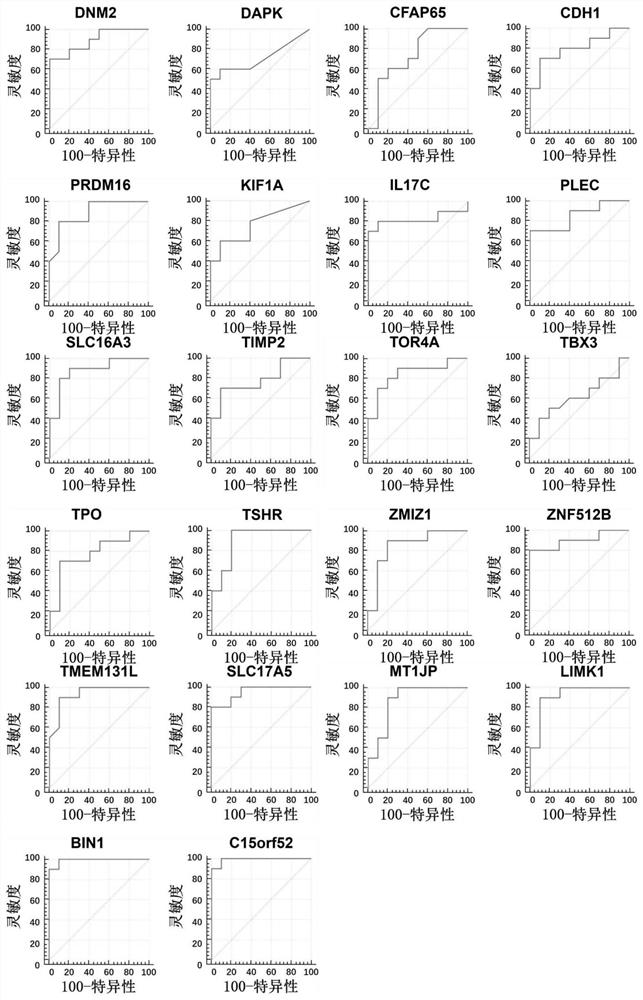
[0427] Example 1: Reduced Representation Bisulfite Sequencing (RRBS) screening of methylation sites for differences between benign and malignant thyroid nodules
[0428] 1) Sample preparation
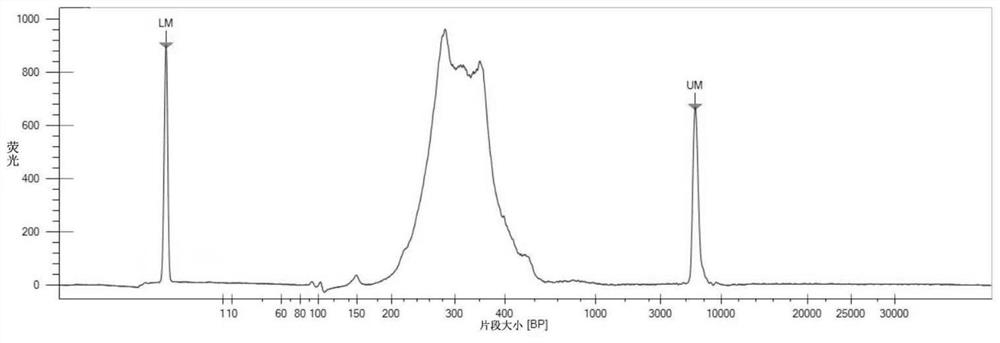
[0429] DNA was extracted from 37 cases of thyroid cancer and 37 cases of benign thyroid nodules using QIAamp DNA Mini Kit (QIAGEN, product number: 51304); DNA concentration was detected by QubitTM dsDNA HS Assay Kit (Thermo, product number: Q32854); 1% agarose gel electrophoresis for quality inspection.
[0430] 2) MspI digestion
[0431] Prepare the reaction system as follows:
[0432]
[0433]
[0434] The reaction procedure is: 37°C for 2 hours, and store at 4°C.
[0435] 3) End repair and A addition
[0436] Prepare the reaction system as follows:
[0437] components Volume (μl) Digested DNA product 20.0 End Repair&A-Tailing Buffer 4.0 End Repair&A-Tailing Enzyme Mix 2.0 nuclease free water 14.0 total 40.0
[0438] The reaction...
Embodiment 2
[0481] Example 2, Methylation-specific PCR (Methylation-specific PCR, MSP) and quantitative methylation-specific PCR (Quantitative MSP, Q-MSP) to verify differential methylation sites
[0482] 1) Sample preparation
[0483] Use the QIAamp DNA Mini Kit (QIAGEN, Cat. No.: 51304) to extract DNA from tissues or plasma of 10 cases of thyroid cancer and 10 cases of benign thyroid nodules; use Qubit TM dsDNA HS Assay Kit (Thermo, catalog number: Q32854) was used to detect the concentration of DNA; 1% agarose gel electrophoresis was used for quality inspection.
[0484] 2) DNA conversion
[0485] Use MethylCode TM Bisulfite Conversion Kit (Thermo, Cat. No.: MECOV50) was used to perform bisulfite conversion on the DNA obtained in step 1, and unmethylated cytosine (cytosine, C) was converted into uracil (uracil, U); The cytosine does not change after conversion.
[0486] 3) PCR mixture preparation
[0487] Including PCR reaction solution, primer mix, probe mix, the preparation of...
Embodiment 3
[0498] Example 3, multiple pre-amplification methylation-specific PCR method (preAMP-MSP) for discrimination of benign and malignant thyroid nodules
[0499] 1) Sample preparation
[0500] Use QIAamp Circulating Nucleic Acid Kit (QIAGEN, catalog number: 55114) to extract cfDNA from the plasma of 20 cases of thyroid cancer and 20 cases of benign thyroid nodules; use Qubit TM dsDNA HS Assay Kit (Thermo, Cat. No.: Q32854) was used to detect the concentration of cfDNA; LabChip 3K Assay was used for quality inspection.
[0501] 2) DNA conversion
[0502] Use MethylCode TM Bisulfite Conversion Kit (Thermo, Cat. No.: MECOV50) performs bisulfite conversion on the cfDNA obtained in step 1, and unmethylated cytosine (cytosine, C) is converted into uracil (uracil, U); The cytosine does not change after conversion.
[0503] 3) Pre-amplification PCR reaction
[0504] The pre-amplification PCR mixture includes PCR reaction solution and primer mixture. The primer mix included RARG (S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com