Epigenetic markers of colorectal cancers and diagnostic methods using the same
A large intestine and coordinate technology, applied in biochemical equipment and methods, microbial measurement/testing, nucleotide library, etc., can solve problems such as expensive, low patient willingness to swallow, invasiveness, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[1475] Bisulfite labeling was employed to generate methylated and unmethylated genomic fractions as described below. Briefly, DNA was digested with the methylation-insensitive enzymes MspI and TaqI and treated with sodium bisulfite under non-denaturing conditions so that if it was unmethylated, the DNA left by each restriction enzyme Cytosines in the 5'-CG single-stranded overhang will be converted to uracil, but will remain unconverted if methylated. Separate adapters with 5'-CG or 5'-CA overhangs were ligated to provide the ligated methylated and unmethylated portions, respectively. After incorporation of the second primer by random priming copies of the reverse strand, this common primer is used in combination with the appropriate forward primer to amplify the methylated and unmethylated fractions.
[1476] In detail, tumor samples and matched normal DNA samples from 8 patients were processed and analyzed as described below.
[1477] 1. Using a Bioruptor UCD-200 sonicator...
Embodiment 2
[1523] Methylated DNA fractions were prepared from bisulfite-treated DNA of colorectal cancer cell lines HCT116, HT29 and SW480 and from DNA isolated from whole blood using the biotin capture method described below , and the library of methylated DNA was sequenced using the Applied Biosystems SOliD sequencing system. Briefly, DNA was cut and a modified SOLiD P2 adapter was ligated to the cut DNA. The DNA was then cleaved with Csp61 (cleavage site G'TAC) and ligated with a modified SOLiD P1 linker. The DNA is then denatured and treated with sodium bisulfite to convert all unmethylated cytosines to uracils. The bisulfite-treated DNA was copied using the modified P2 primer, and the original uracil-containing bisulfite-treated DNA strand was removed. The P1 forward primer was then used to prime forward strand synthesis in the presence of biotin-dCTP. Biotin dCTP is thus incorporated at positions in the original DNA that contained methylated cytosines and were thus not converted...
Embodiment 3
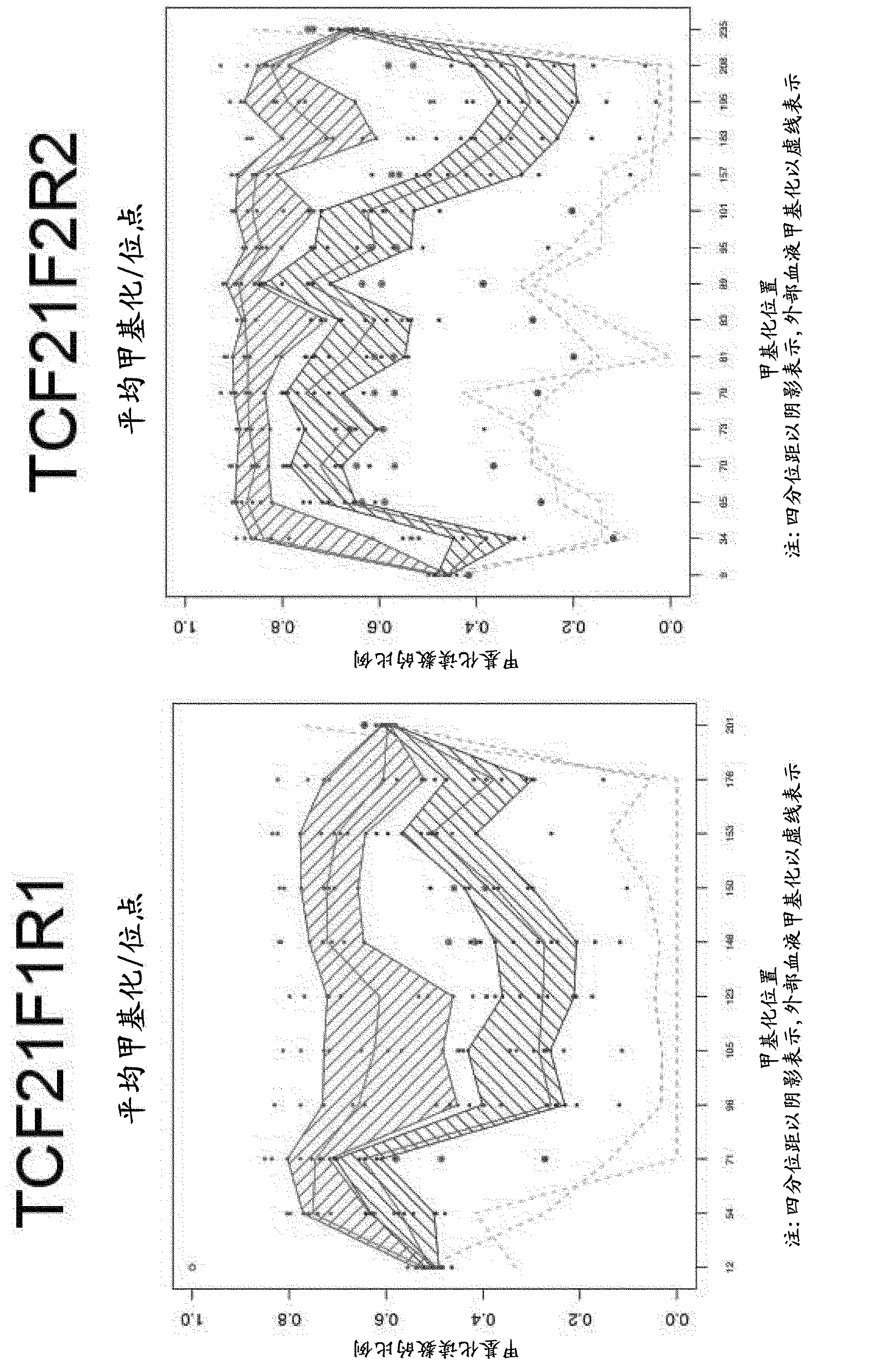
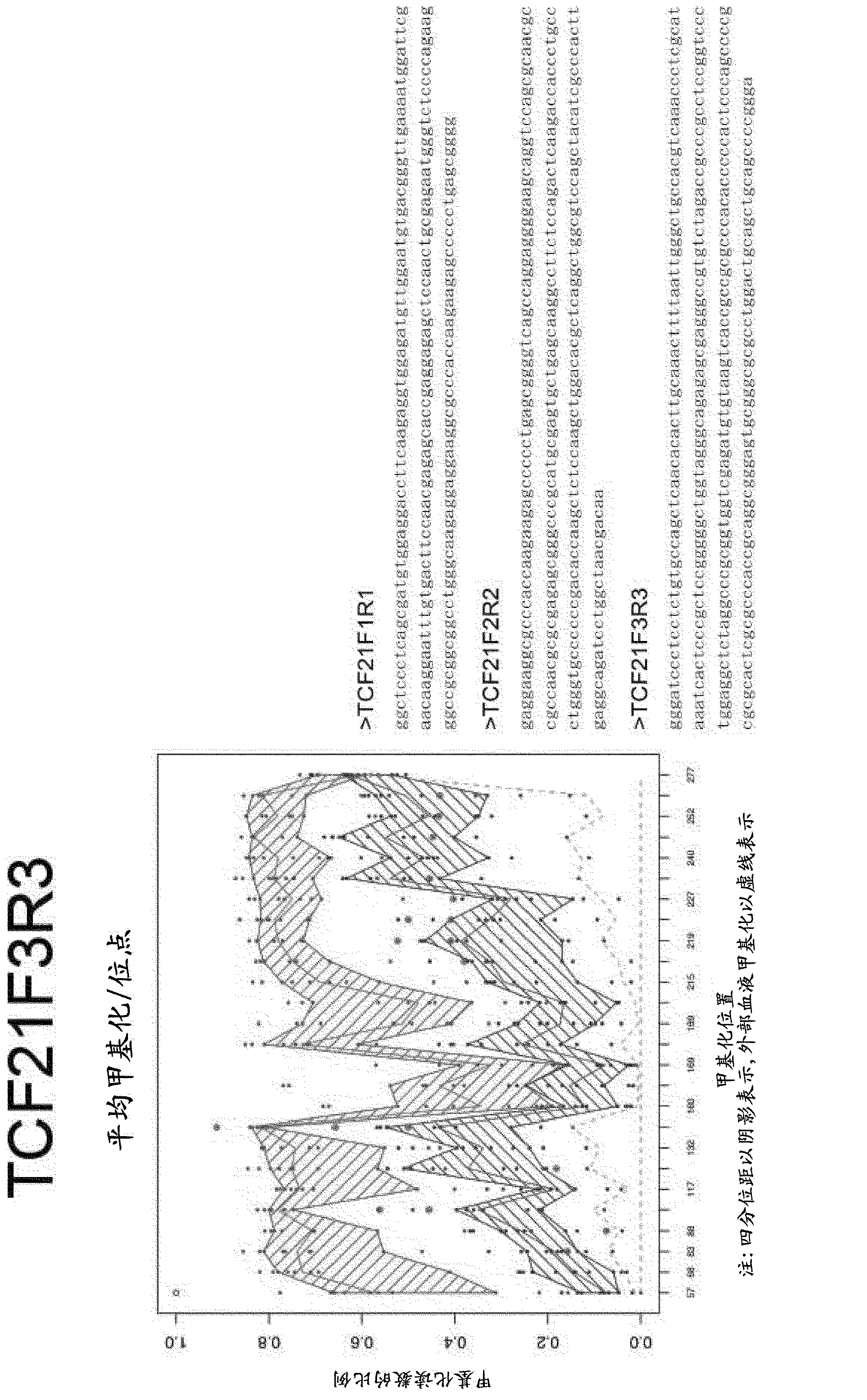
[1550] DNA methylation profiles of selected genes in colorectal cancer DNA and normal tissue DNA
[1551] Primers were designed for methylation status-independent amplification of promoter regions of genes and / or gene sets identified in previous examples. Gene coordinates, primer coordinates and chromosome coordinates of the amplicons are shown in Table 5.
[1552] Primers were used for PCR from 10 colorectal cancer specimens, their bisulfite-treated DNA from matched normal tissues, and normal blood DNA. Use Promega GoTaq master mix (without SybrGreen), 4mM MgCl 2 Amplification was performed with 200 nM primers and 10 ng input DNA. Cycling conditions were 95°C for 2 minutes (1 cycle), followed by 50 cycles of the following: 95°C for 15 seconds; N°C for 30 seconds; 72°C for 30 seconds, where the annealing temperature N for each amplicon is shown in Table 5. For some amplicons, an additional 200 μM dATP and dTTP were added to enable comparable amplification of methylated and un...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com