Aptamer for specific recognition of amantadine and its application
An amantadine and aptamer technology, which is applied in the field of aptamers that specifically recognize amantadine, can solve the problems of natural structure damage and lack of the target, and achieves the effects of high sensitivity, high affinity and easy preparation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Synthesis of aptamers and capture probes
[0027] Aptamer sequences with random libraries and 5' biotinylated capture probes were synthesized by Shanghai Bioengineering Co., Ltd.
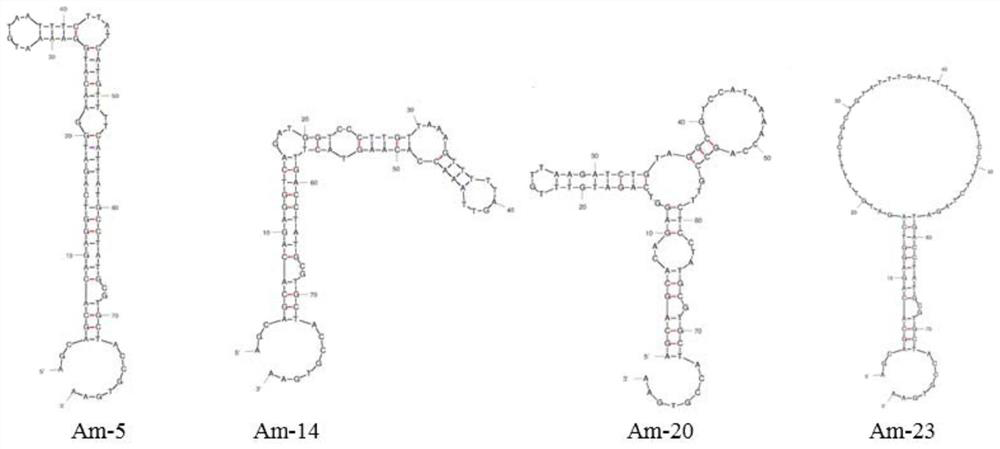
[0028] Aptamer sequence (SEQ ID NO.2):
[0029] 5'-AGCAGCACAGAGGTCAGATG-N40-CCTATGCGTGCTACCGTGAA-3'
[0030] Biotinylated capture probe (SEQ ID NO.3):
[0031] 5'-botin-GACCCTCTGTGCTGCT-3'.
Embodiment 2
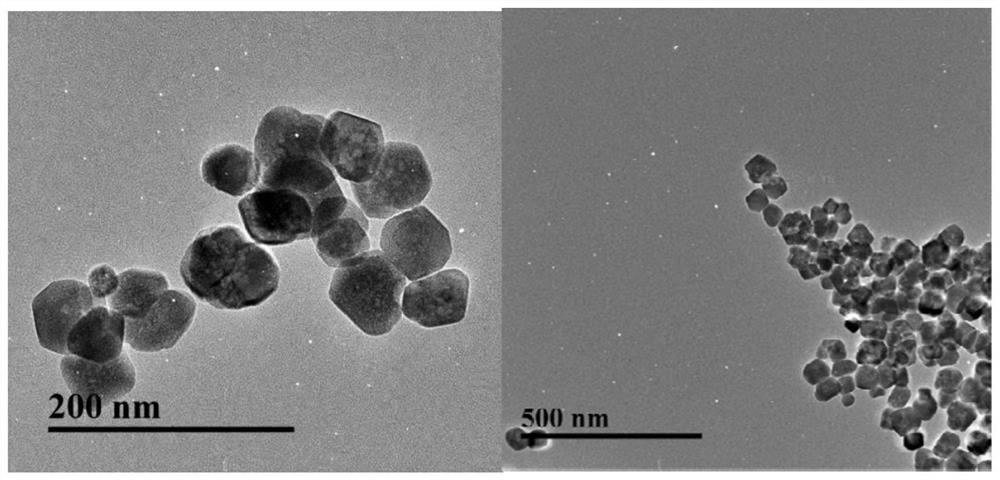
[0032] Example 2: Fe 3 o 4 Preparation and coating of magnetic nanoparticles
[0033] 2.1 Preparation of aminated magnetic beads
[0034] One-step preparation of aminated magnetic beads using hot melt, the process is as follows: 1. Add 30mL of ethylene glycol, 1g of ferric chloride hexahydrate and 2g of sodium acetate into a round bottom flask in turn, shake well and add 6.50g of 1,6- Hexamethylenediamine; 2. Magnetically stir the above solution in a 50°C water bath until a uniform purple-red colloidal solution; 3. Transfer the solution to a 100mL polytetrafluoroethylene-lined autoclave, and heat it at 198°C 6h; 4. After the reaction kettle is cooled, remove the supernatant in the kettle under the action of an external magnetic field, add 100mL absolute ethanol to the kettle, stir and pour it into a beaker; 5. Remove the supernatant by magnetic separation, and then add 100mL of water for ultrasonic cleaning and magnetic separation. According to this method, wash three time...
Embodiment 3
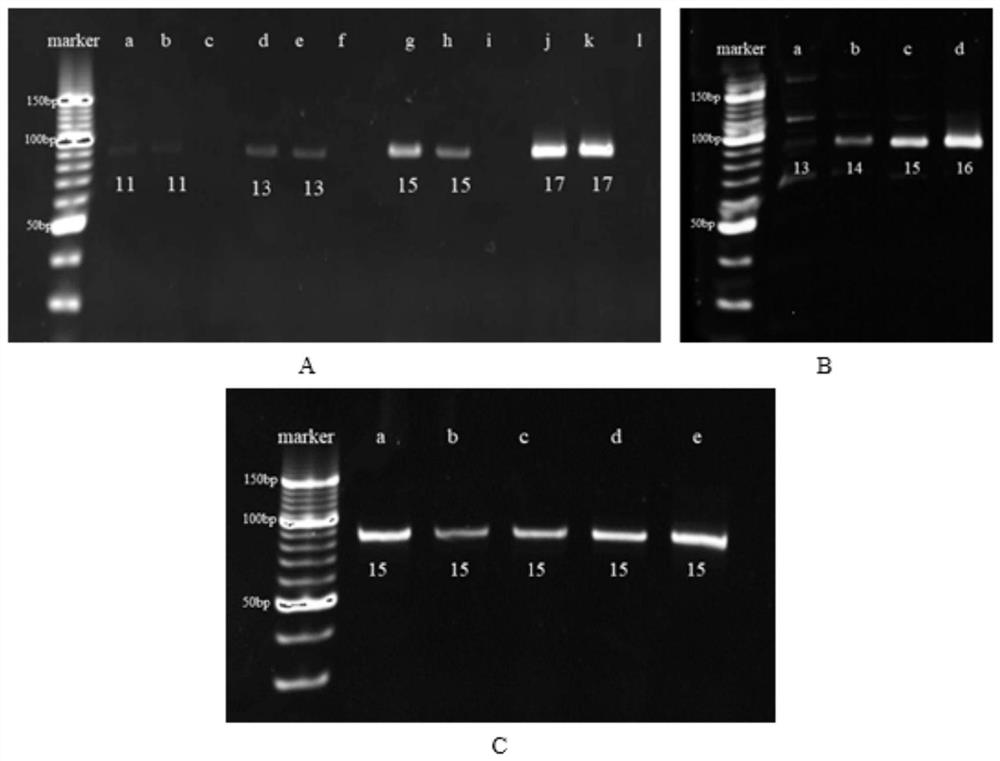
[0037] Example 3: Screening of amantadine aptamer sequences
[0038] 3.1 Library fixation and screening
[0039] The capture-SELEX technology based on magnetic separation is used for circular screening, and the steps are as follows: before each round of screening, the library or secondary library is combined with the biotinylated capture probe at a molar ratio of 1:2, and a certain amount of After denaturation at 95°C for 10 minutes, incubate in a 37°C, 200rpm incubator for 2h; after the incubation, add avidinated aminated magnetic beads and incubate in an incubator at 37°C, 200rpm for 3h. Using the specific binding of biotin and avidin, the library or secondary library is immobilized on magnetic beads. After the incubation, the supernatant was kept, and the immobilization degree of the library was verified by UV spectrophotometry; the magnetic beads after the immobilized library were washed 5 times with a buffer solution, and the target amantadine was dispersed in the buffer...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com