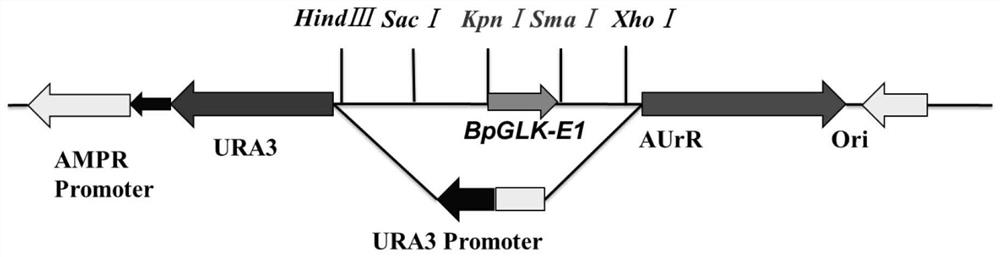
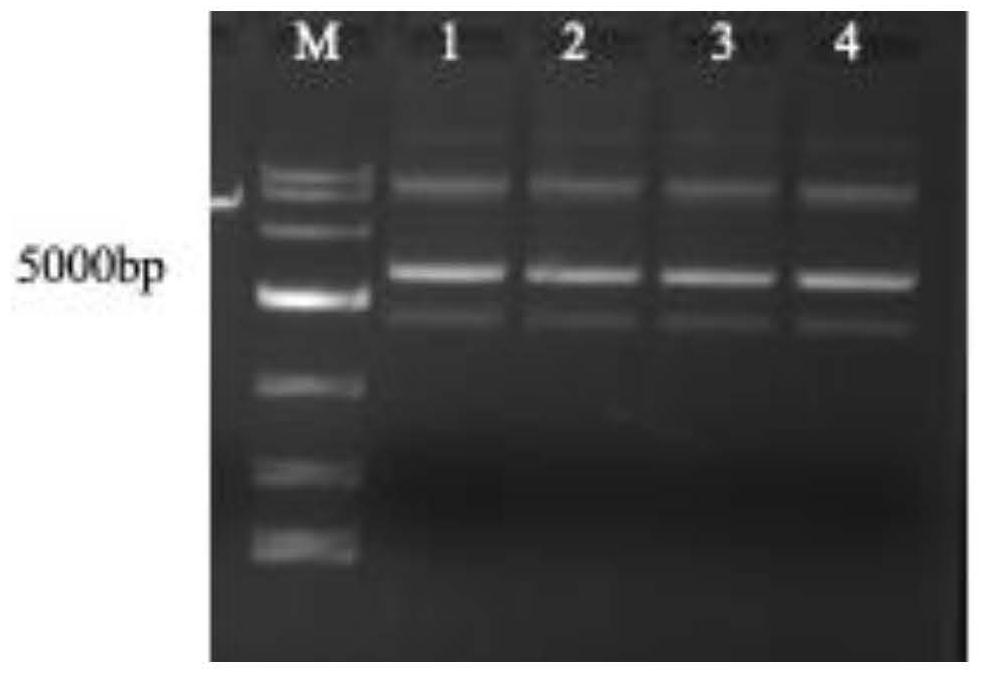
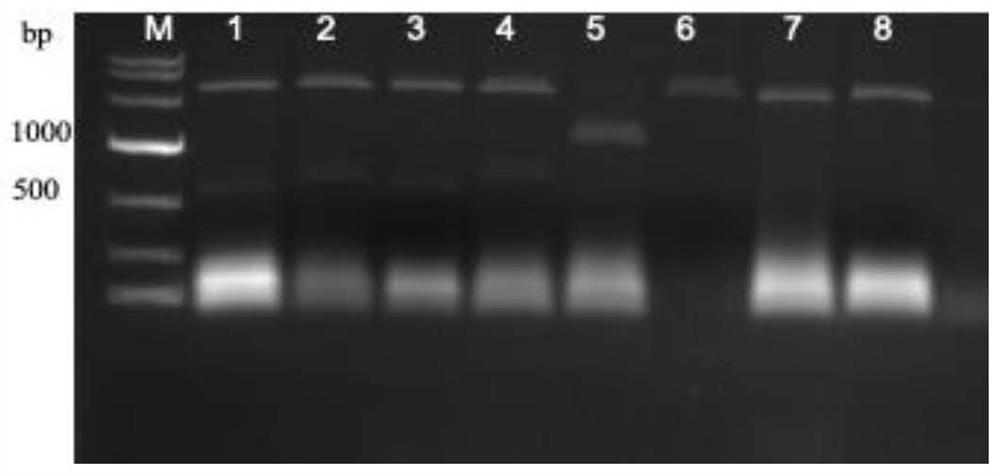
CRISPR/Cas9 system for targeted knockout of GLK gene and application of CRISPR/Cas9 system
A genetic and targeting technology, applied in the field of molecular biology, can solve the problems of long required period and achieve the effect of enriching varieties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1B
[0027] Example 1 Prediction of pGLK gene target site and detection of gRNA activity
[0028] 1. Prediction of BpGLK gene target site and gRNA sequence design
[0029] According to the gene sequence and coding region sequence of birch BpGLK (Bpev01.c0167.g0013.m0001), the CRISPR-direct web page (http: / / crispr.dbcls.jp / ) was used to screen for specific targets. That is, find NGG (N is A, T, C or G) in the CDS region sequence of the gene, preferably AGG, and take the 20 bp in front of NGG as the target sequence in order to improve the gene knockout efficiency. Design gRNA according to the target sequence, see Table 1 for details.
[0030] Table 1 gRNA sequence
[0031]
[0032] 2. Synthesis and activity detection of gRNA
[0033] Add the T7 promoter sequence at the front end of the gRNA sequence: TAATACGACTCACTATAG; add the guide sequence GTTTAAGAGCTATGCTGGAAACAGCATAGCAAGTTTAAATAAGG (stem-loop structure) at the back end of the gRNA sequence; use the primers for gRNA synthes...
Embodiment 2
[0053] Example 2 BpGLK gene editing mediated by gene gun
[0054] 1. Preparation of gene gun particles (the following operation steps can bombard 60 guns according to the amount of 500 μg per gun)
[0055] (a) Add 30mg of 1.0μm Gold Microcarriers into Eppendorf tubes.
[0056] (b) Add 1ml of 70% alcohol, vortex fully, let stand for 15min, and centrifuge at 1500rpm for 5min.
[0057] (c) Discard the supernatant, add 1ml of sterile water, vortex well, centrifuge at 1500rpm for 5min, and repeat 3 times.
[0058] (d) Add 1 ml of 50% glycerin, and the final concentration reaches 60 mg / ml at this time, and store at 4°C.
[0059] 2. Package of CRISPR / Cas9 (RNP) (the amount below is enough for 6 shots, adjust the amount as needed)
[0060] (a) Vortex the above-prepared Gold Microcarriers mother solution for 5 minutes to break up agglomerated particles. It should be noted that the particles are easy to sink, and the resuspended particles must be re-oscillated every time the gun is ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com