CRISPR/Cpf1 vector suitable for deep-sea fungi FS140 as well as construction method and application of CRISPR/Cpf1 vector
A construction method, cpf1 technology, applied in the direction of microorganism-based methods, introduction of foreign genetic material using vectors, application, etc., can solve problems such as gene knockout of deep-sea fungi that have not been seen yet
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
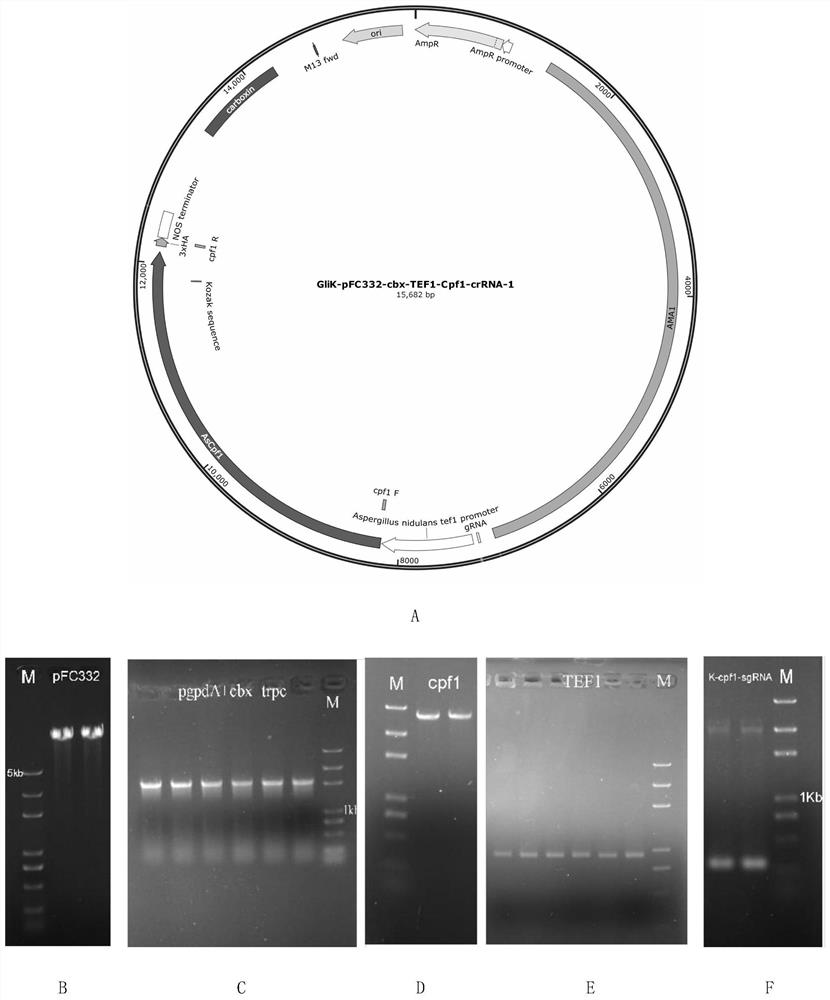
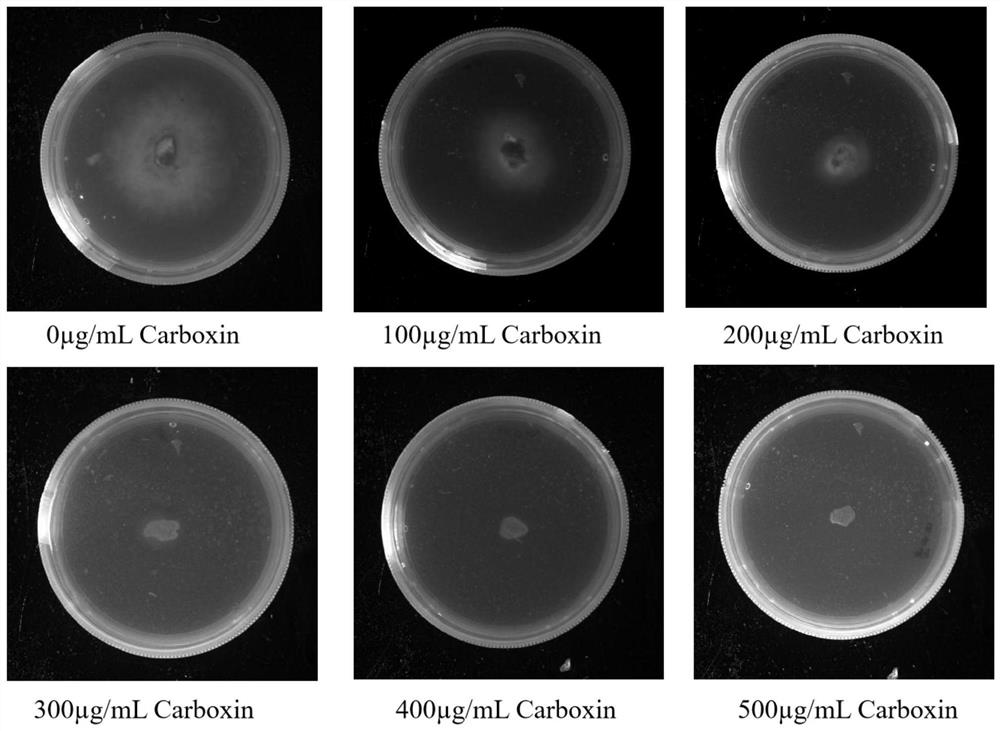
[0042] Example 1: Construction of targeted gliotoxin biosynthesis gene knockout vector
[0043] On the basis of the pFC332 plasmid, pFC332 was transformed, and PacI and BglII sites were used for double enzyme digestion, and the hygromycin resistance gene was replaced by the wisterin resistance gene; the Cpf1 protein was replaced by Cas9; a sequence was inserted: SNR52 Promoter (BglII): crRNA: SUP4 terminator (PacI) (which contains the SNR52 promoter, crRNA targeting sequence backbone gene and its terminator), enabling the plasmid to stably express Cpf1 protein and transcribe crRNA simultaneously after transformation into protoplasts (The plasmid was named pFC332-cbx-TEF1-Cpf1-crRNA).
[0044] The primers designed for the construction of the vector are shown in Table 1, and the GliK-pFC332-cbx-TEF1-Cpf1-crRNA-1 vector was constructed by restriction enzyme ligation and homologous recombination. The construction method is as follows:
[0045] Table 1 Primer Sequence
[0046] ...
Embodiment 2
[0053]Example 2 The method for introducing exogenous genes into G.pallida FS140 protoplasts is as follows:
[0054] 1) Pick an appropriate amount of G. pallida FS140 mycelium and inoculate it in 200 mL of PDB liquid medium, and culture it at 30° C. and 180 r / min for 3-4 days. Select 2 g (wet weight) of well-grown bacterium balls in a 50 mL centrifuge tube and wash twice with PBS buffer to fully wash away the residual PDB medium. Weigh 0.15g lyase and dissolve in 20mL KC buffer (0.6M KCl, 0.05MCaCl 2 ), and filtered with a 0.22 μm filter membrane, added to the washed bacterial balls, and placed in a shaker at 28°C and 80r / min for about 3 hours to lyse. Filter the lysate with a 200-mesh filter, filter the hyphae, and then filter again with 6 layers of lens tissue, centrifuge at 4°C and 6500×g for 10 minutes, and discard the supernatant. Add 5mL of KC buffer, gently blow and mix the precipitate with a pipette gun, centrifuge at 4000×g for 5min at 4°C, and discard the supernatan...
Embodiment 3
[0060] 将Glik基因(核苷酸序列为:atgaccatacaactccctcacacacctgacagagaagaaggccctggtgc ttctcctgcatgcaagttcaatgcaatccagacattccggtggttatgggacctagtcatcccaactagcgaccttactc aagaaaccggtcgatatcctccgaagacgacgatcgagagacggcgcgcatcaaccacagatagatcgctcgataaggac gaatacctagctgagaaggtcgcaggtcatcaggtcgaagaacatgtccccccggagaagaccgtcctctacctcgcgta cggctcgaatctggccgcggagaccttcctgggcaagcgaggcattaggcctctatcccagatcaacgttgtcgttcccg gcctacgactaactttcgaccttcctgggttaccatacgttgagccatgcttcgcagcgactcggcactggactcataca ccaagagtaacacaaacagaaggaaatggaagtaatgaaggggtcgatgcagaagtgttggagaattcgtccctcgtgcc acaggagaagaacgacatgcctctcgtcggcgtcgtgtatgaggtcactgtcgccgattatgccaagataatagccacag agggtggtgggcgcggataccgagacgtcgttgtcgattgctatccttttcccaaatcatacagtcccaccgatccggtc cctgagtgccccgaaaccaaacccttcaagtcccacaccctcctctctccagccgacgacgcagtctcgggtctcctggc cgcagggaagtcataccgacccgtccgacccaatcctggctacgcccagccctccgcttga)插入到pET-30a载体的酶切位点NdeI和XhoI之间的位置,获得pET30a-GliK,转化进大肠杆菌 BL21(DE3) competent cells were screened for positive clones to obtain Escheri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com