A kind of human papillomavirus type 35/hpv35 type l1/l2 and its preparation and application
A technology of human papillomavirus and pseudovirus, which is applied in the biological field to achieve the effects of simple interpretation, short detection cycle and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 Optimization of HPV 35L1 protein and HPV 35L2 protein coding gene
[0027] The genes encoding HPV 35L1 protein and HPV35L2 protein were optimized based on multiple dimensions such as codon usage frequency, RNA secondary structure, and GC content. The amino acid sequence of HPV 35L1 protein is shown in SEQ ID NO.1 (GenBank accession number: CAA52566.1), and the amino acid sequence of HPV 35L2 protein is shown in SEQ ID NO.2 (GenBank accession number: CAA52565.1). The gene encoding the HPV 35L1 protein before optimization is SEQ ID NO.3, and the gene encoding the HPV 35L1 protein after optimization is SEQ ID NO.4 (HPV 35L1-op1) and SEQ ID NO.5 (HPV 35L1-op2). The gene encoding the HPV35L2 protein before optimization is SEQ ID NO.6, and the gene encoding the HPV 35L2 protein after optimization is SEQ ID NO.7 (HPV 35L2-op1) and SEQ ID NO.8 (HPV 35L2-op2).
[0028] SEQ ID NO.1 (amino acid sequence of HPV 35L1 protein)
[0029]MSLWRSNEATVYLPPVSVSKVVSTDEYVTRTNIYYHA...
Embodiment 2
[0044] Construction and preparation of embodiment 2 HPV 35 type pseudoviruses
[0045] Step 1: Insert HPV 35L1, HPV 35L1-op1, HPV 35L1-op2, HPV 35L2, HPV 35L2-op1, HPV35L2-op2, and EGFP (fluorescent protein reporter) genes into pCDNA3.1 plasmids to obtain expression plasmids 35L1-pCDNA3, respectively .1 (SEQ ID NO.9), 35L1-op1-pCDNA3.1 (SEQ ID NO.10), 35L1-op2-pCDNA3.1 (SEQ ID NO.11), 35L2-pCDNA3.1 (SEQ ID NO.12 ), 35L2-op1-pCDNA3.1 (SEQ ID NO.13), 35L2-op2-pCDNA3.1 (SEQ ID NO.14), EGFP-pCDNA3.1 (SEQ ID NO.15).
[0046] Expression plasmid 35L1-pCDNA3.1 sequence (including pre-optimized gene HPV 35L1), SEQ ID NO.9:
[0047]GACGGATCGGGAGATCTCCCGATCCCCTATGGTGCACTCTCAGTACAATCTGCTCTGATG CCGCATAGTTAAGCCAGTATCTGCTCCCTGCTTGTGTGTTGGAGGTCGCTGAGTAGTGCG CGAGCAAAATTTAAGCTACAACAAGGCAAGGCTTGACCGACAATTGCATGAAGAATCTG CTTAGGGTTAGGCGTTTTGCGCTGCTTCGCGATGTACGGGCCAGATATACGCGTTGACAT TGATTATTGACTAGTTATTAATAGTAATCAATTACGGGGTCATTAGTTCATAGCCCATATAT GGAGTTCCGCGTTACATAACTTACGGTAAATGGCCCGCCTGGCTGACCGCCCAA...
Embodiment 3
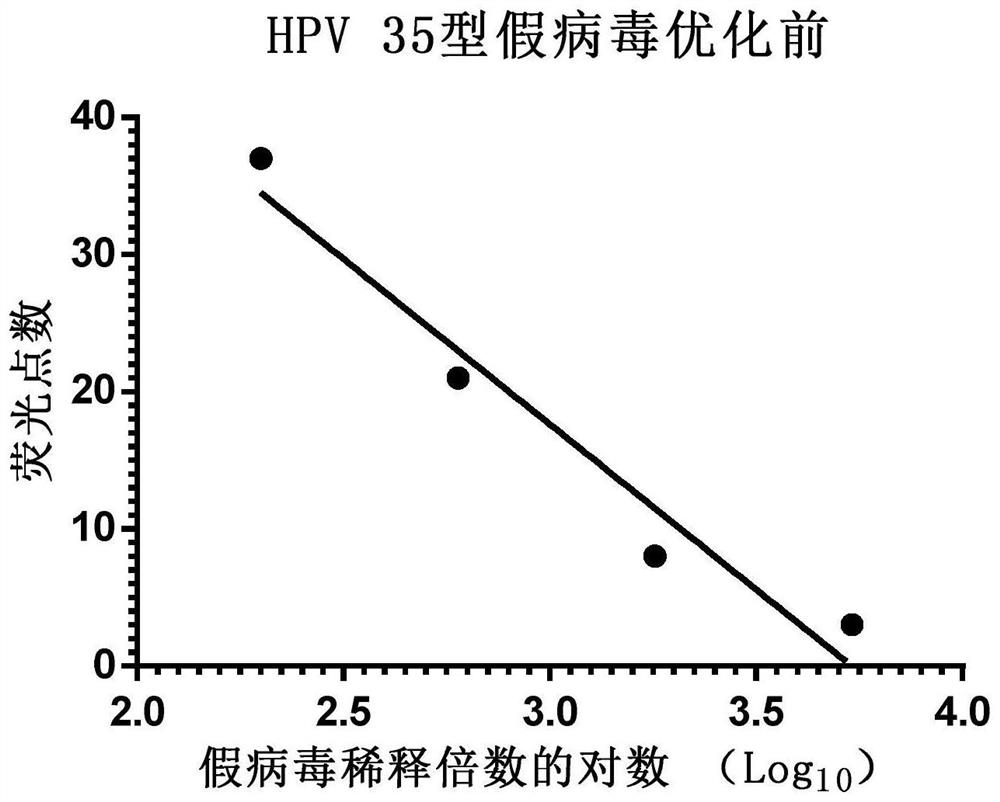
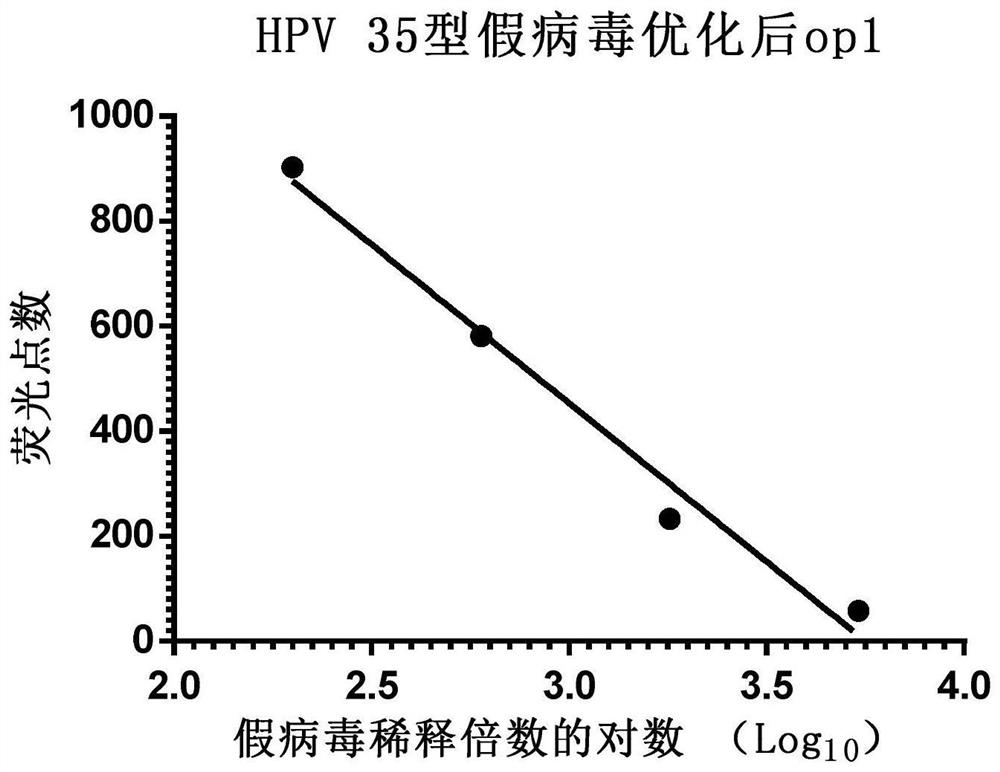
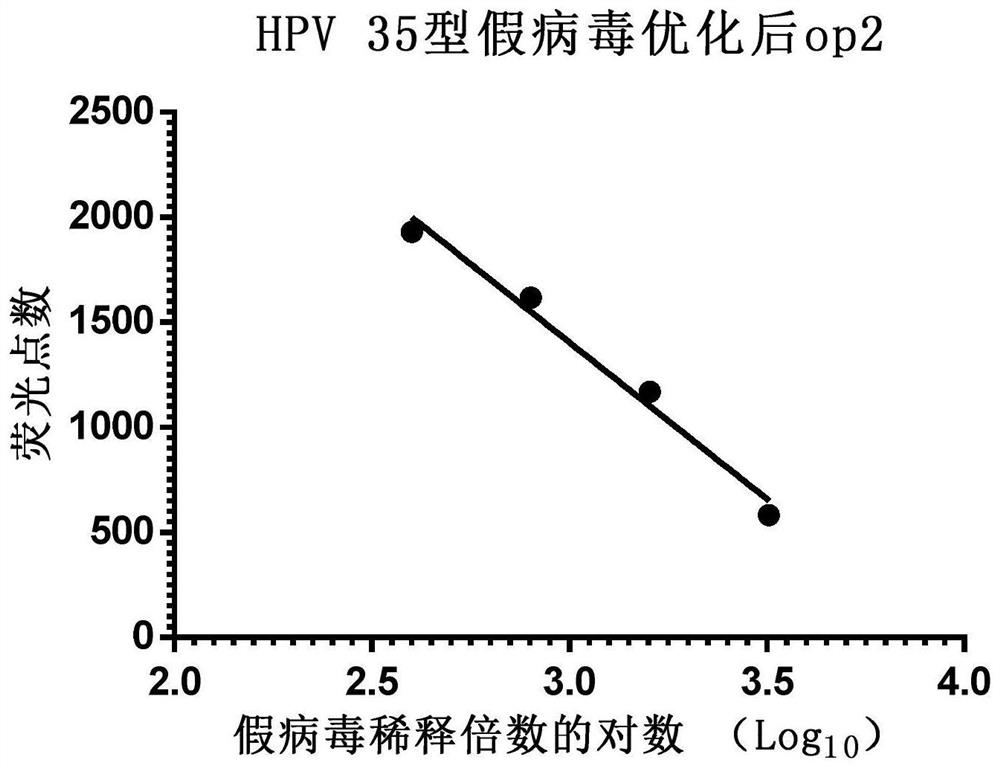
[0069] Example 3 Application of Enzyme-Linked Fluorescent Spot Analyzer to Detect HPV Type 35 Pseudovirus Using Multiples
[0070] Step 1: Collect 293FT cells in the logarithmic growth phase, count with a hemocytometer and dilute the cells with DMEM complete medium to a density of 1.5×10 5 pieces / ml.
[0071] Step 2: In a 96-well cell culture plate, add 150 μl of 293FT cells per well, place at 37°C, 5% CO 2 Cultivate in an incubator for 16-24 hours.
[0072] Step 3: Take the pseudovirus solution and dilute it with DMEM complete medium to 200 times or an appropriate multiple, then dilute according to the 2-fold ratio, and serially dilute 4 to 8 dilutions.
[0073] Step 4: Add 50 μl pseudovirus dilution solution to each well, set 8 wells to repeat for each dilution, and set a blank control at the same time, the blank control is 50 μl DMEM complete medium, after culturing for 72 hours, use an enzyme-linked fluorescence spot analyzer to detect each well of green fluorescent dot...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com