Targeting capture probe of tumor fusion gene and application thereof
A fusion gene and targeted capture technology, which is applied in the determination/testing of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problem of not being able to clarify the detailed break site of the gene, and not publishing the principle of probe design and probe sequence , No fusion gene detection and other problems have been found, which is conducive to remote inspection, fast detection time, and improved detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
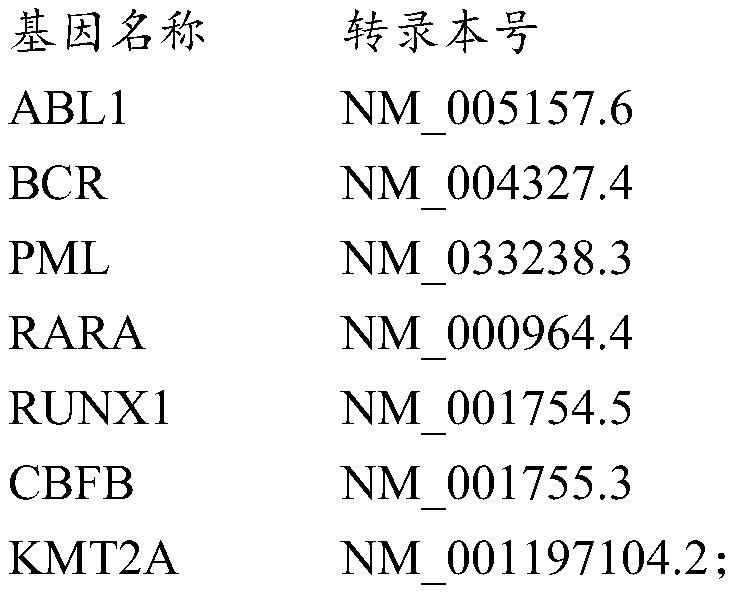
Image
Examples
Embodiment 1
[0093] Taking the probe sequence SEQ ID NO: 312 of exon 2 of the RARA gene exon: CGGTGCCTCCCTACGCCTTCTTCTTCCCCCCTATGCTGGGTGGACTCTCCCCGCCAGG CGCTCTGACCACTCTCCAGCACCAGCTTCCAGTTAGTGGATATAGCACACCATCCCCA GCCA as an example, after comparing the sequences to the human transcription probe database at http: / / grch37.ensembl.org / Multi / Tools / Blast, the results show that, The 100% consensus sequence of the needle is the transcript level sequence of the RARA gene, and only 17 bases overlap with the other gene sequences at most, indicating that the probe has good specificity.
Embodiment 2B
[0094] Example 2 Detection of BCR-ABL1 fusion gene
[0095] Specimen information: Bone marrow specimens from patients with positive BCR-ABL1 fusion gene
[0096] Experimental steps:
[0097] (1) Extract total RNA from bone marrow specimens and use Qubit TM The RNA IQ Assay Kit detects the integrity of the sample RNA, and the integrity of the RNA (RIN value) is required to be greater than or equal to 6.5. The initial total amount of total RNA is required to be in the range of 1-100ng, and the RNA is dissolved in 10 microliters of nuclease-free water.
[0098] (2) Resuspend the starting RNA in Fragmentation, Prep and Elution Buffer (1X), and fragment it into the desired fragment size by incubating at high temperature. The size of the target fragment we need is about 400bp, and the fragmentation conditions are: 85°C, 6 minutes.
[0099] (3) RNA is reversed into cDNA (two-step method), and the first strand is synthesized by random primer combination. When synthesizing the sec...
Embodiment 2
[0110] Implementation of 2 rare BCR-ABL1 fusion gene detection
[0111] Specimen information: bone marrow specimens from patients with positive BCR-ABL1 fusion gene.
[0112] Experimental procedure: with embodiment 2.
[0113]Detection instrument: illumina nextseq550.
[0114] Detection results: BCR-ABL1 (e8a2 type) fusion gene transcripts were detected.
[0115] The analysis results of STAR-FUSION software are shown in Table 2.
[0116] Table 2 BCR-ABL1 (e8a2 type) transcript
[0117]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com