Formic acid and CO2 autotrophic recombinant escherichia coli and construction method thereof
A technology for recombinant Escherichia coli and construction methods, applied in the directions of botanical equipment and methods, microorganism-based methods, biochemical equipment and methods, etc., can solve the problems of poor general performance, long operation cycle, high input cost, and achieve universality The effect of high, short cycle time and simple construction process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
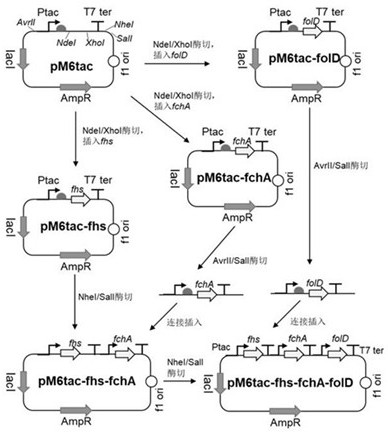
[0039] Example 1: Modular plasmid construction of formic acid assimilation function
[0040] According to the gene sequence and map of the commercial plasmid pETDuet-1, the upstream primer pEM6-F carrying the SalI restriction site and the downstream primer pEM6-R carrying the AvrII restriction site were respectively designed, and the pETDuet-1 plasmid was obtained by PCR amplification The linearized fragment; the plasmid linearized fragment obtained by SalI / AvrII double digestion and the tac gene synthesis fragment (sequence is the sequence between AvrII and SalI restriction sites shown in SEQ ID NO.4), passed through T4 DNA ligase was used to ligate the digested plasmid linearized fragment and tac gene fragment at 16°C for 1 hour to transform Escherichia coli JM109 competent; finally, the plasmid containing pEM6tac ( SEQ ID NO.4 is the clone of the plasmid sequence), amplified and extracted for later use. According to Clostridium ljungdahlii ( Clostridium ljungdahlii ) of ...
Embodiment 2
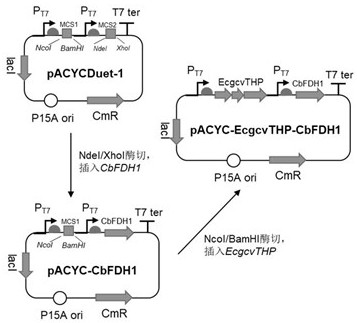
[0043] Example 2: CO 2 Modular plasmid construction of assimilative function
[0044] The glycine lyase complex is generally considered to have a reverse catalytic function under the condition of sufficient reducing cofactor NAD(P)H, that is, to convert CO 2 , NH 4 + and formic acid assimilation product CH 2 - The ability of THF (methylenetetrahydrofolate) to convert to glycine. Therefore, the glycine lyase complex for CO 2 Assimilation plays a key role and is the rate-limiting step in achieving autotrophy in E. coli-dependent C1-pyruvate synthesis pathway.
[0045] According to the glycine lyase of Escherichia coli BL21(DE3) itself EcgcvTHP Gene cluster sequence (as shown in SEQ ID NO.3), respectively design gcvT-F and gcvP-R primers carrying NcoI / BamHI restriction sites (see Table 1), perform PCR amplification on the above sequence to obtain about 4500 bp Gene fragments; use restriction endonucleases NcoI / BamHI to double-enzyme digest the obtained PCR product and pAC...
Embodiment 3
[0048] Embodiment 3: the construction of recombinant escherichia coli BL21 (DE3)
[0049] Escherichia coli BL21(DE3) was used as the chassis bacteria, as the formic acid assimilation plasmid and CO 2 The transformation object of the assimilation plasmid. The preparation method of Escherichia coli competent refers to the routine molecular biology operation steps: first, use LB medium to culture cells at 37°C to a concentration of 0.4 OD 600 ; Secondly, the cells were collected under centrifugation conditions at 4°C and 4000 rpm, and then washed twice with de-cooled sterile ionized water; finally, diluted Escherichia coli with cooled sterile deionized water according to 1000:1, obtained for Transform competent cells with modular plasmids.
[0050] Take 100 ng of pEM6tac-fchA-fhs-folD and pACYC-EcgcvTHP-CbFDH1 plasmids, respectively, and add them to an electric shock cup containing 100 µL of freshly prepared competent Escherichia coli BL21(DE3), mix gently, and incubate on ice ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com