Methods for predicting drug responsiveness in cancer patients
A cancer and pharmacy technology that can be used in biochemical equipment and methods, drug combinations, drug delivery, etc., to solve problems such as missing critical opportunities
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0556] Example 1. Identification of biomarkers of sensitivity and resistance to 2X-121.
[0557] A panel of the NCI60 cell line can be used to determine sensitivity to 2X-121, but other cell line panels can also be used. Here we used the panel of 74 cell lines shown in Table 1.
[0558] Table 1. Measured growth inhibition (IC50 in [mu]M) of cell lines treated with 2X-121.
[0559]
[0560]
[0561]
[0562]
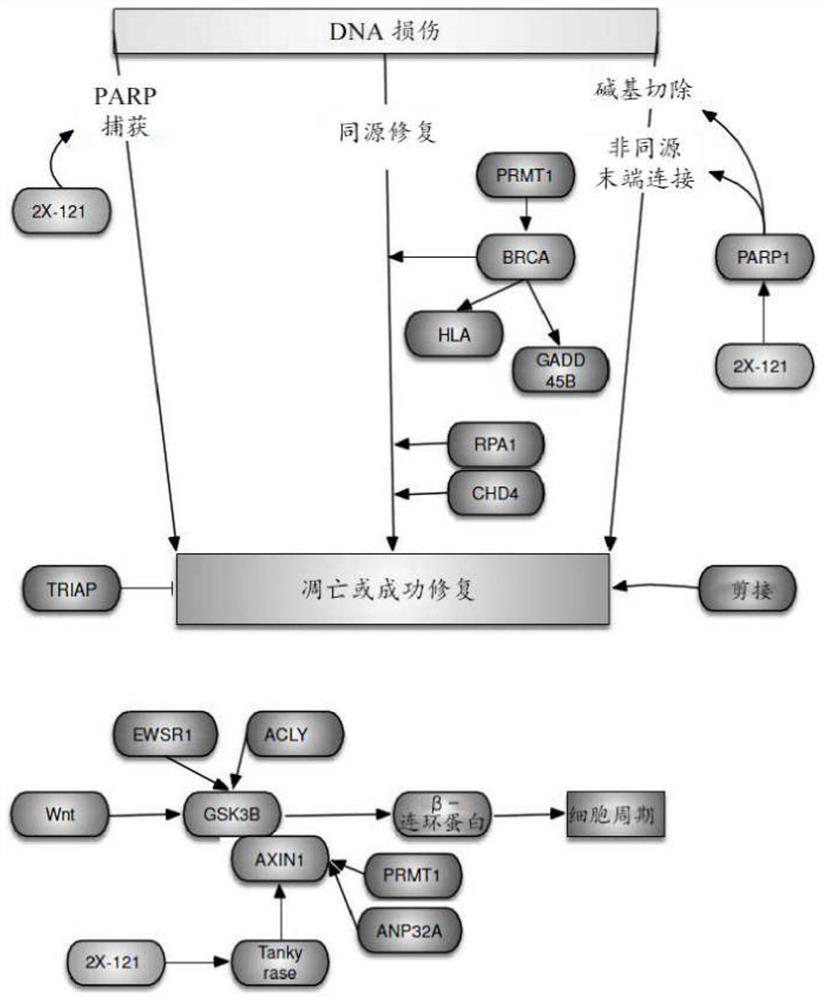
[0563] The 61 cancer cell lines in Table 1 (excluding M14 HEC-1 ECC-1 SNG-11 HEC-88nu SR IM-9 Namalwa MC / CAR U266 JVM-13NCEB- 1 ARH-77) baseline DNA chip. For each array, a logit transformation was performed followed by a Z-transformation so that the mean was zero, SD was 1, and the logit transformation was associated with growth inhibition (-log(GI50)). The expression level of each gene of Table 2 and Table 3 in each cell line correlated with the growth (log(GI50)) of these cell lines in the presence of 2X-121. Pearson correlation coefficients were determ...
example 2
[0583] Example 2. Clinical proof-of-concept study.
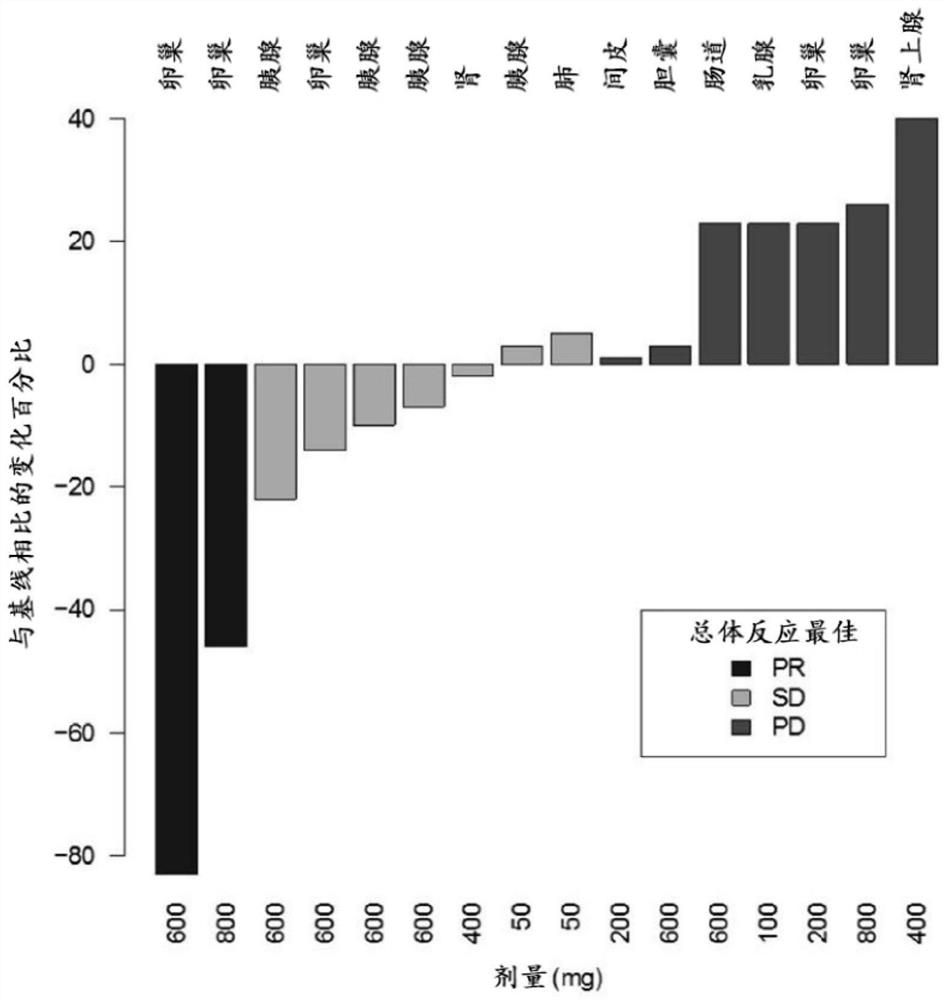
[0584] A phase I trial of 2X-121 was conducted in the United Kingdom (clinicaltrial.gov number NCT01618136). The daily oral dose of 2X-121 is 50-800 mg. Of the 41 patients enrolled in the Phase I clinical trial, 35 were evaluated for response. Among them, 2 cases had partial response (PR) (overall response rate (ORR) 5%), and 13 cases had stable disease (SD). Biopsy and BRCA analysis were voluntary and available from 16 and 7 patients, respectively. figure 2 is a waterfall plot showing the change from baseline tumor diameter for 16 patients who underwent biopsy.
[0585] Of the 16 biopsy patients, 13 passed quality control (QC) in our laboratory and were assayed on the Affymetrix HG-U133Plus2 array. Table 4 shows 13 patients.
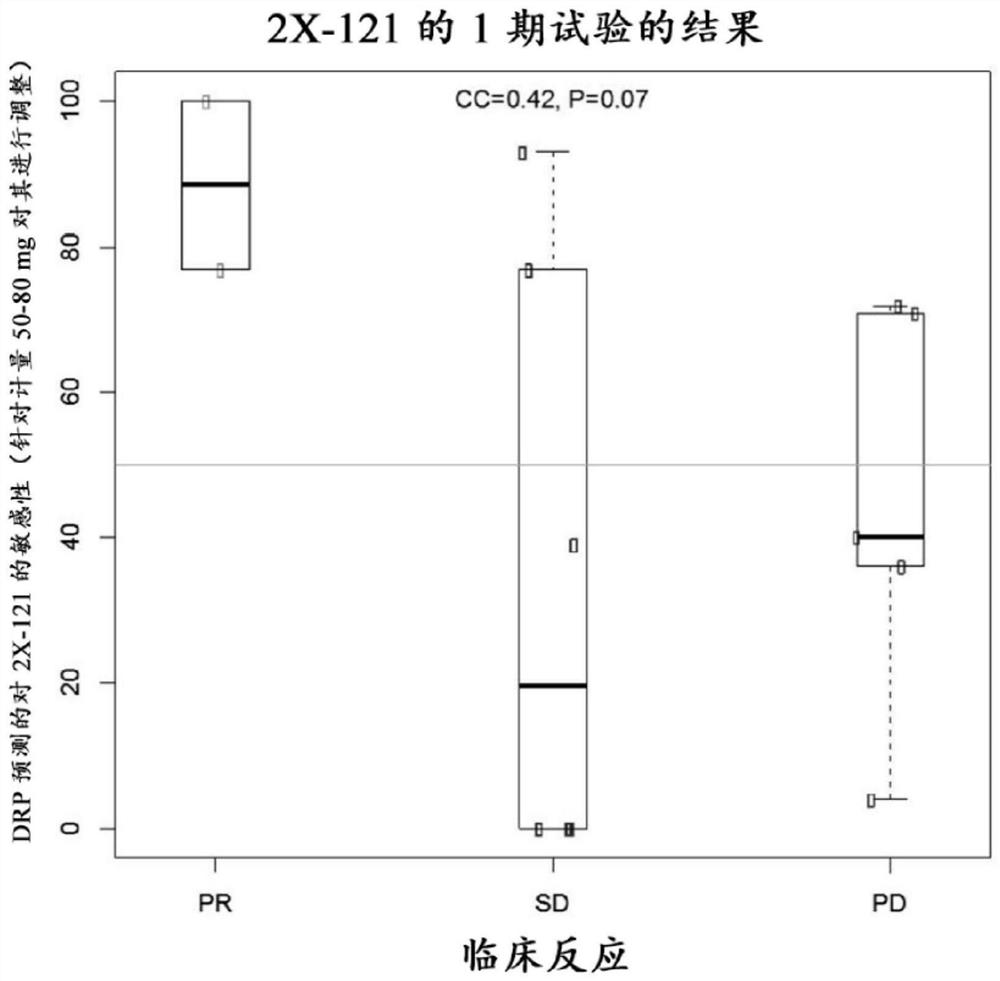
[0586] A statistical analysis plan was completed prior to blinded prediction of 2X-121 sensitivity on 13 samples. In this analysis, for each patient, the difference in the mean expression of th...
example 3
[0609] Example 3. Predicting breast cancer patient responsiveness to 2X-121 or a pharmaceutically acceptable salt thereof.
[0610] The diagnostic methods described herein can be used to predict the responsiveness of breast cancer patients to treatment with 2X-121 or a pharmaceutically acceptable salt thereof. Specifically, the breast cancer patient may be a patient who has not received any cancer treatment before, or a patient who has received cancer treatment other than 2X-121 or a pharmaceutically acceptable salt thereof. Still further, the patient can be a patient diagnosed with breast cancer or suffering from recurrence of breast cancer.
[0611] A biological sample (eg, a breast tissue sample such as obtained by biopsy) can be obtained from a patient by methods well known in the art. The sample can be frozen and / or prepared by, for example, formalin fixation and paraffin embedding. Specifically, for one or more of the biomarkers shown in Table 2 and / or Table 3, mRNA ca...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com