Prognostic biomarkers and detection kits for patients with intrahepatic cholangiocarcinoma
A technology for detecting reagents and inner bile ducts, which is applied in the field of biomedicine, can solve the problems of complex evaluation calculation methods and limited consistency of evaluation results, and achieve the effects of stable prediction, high accuracy, and strong reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0095] Example 1: Detection of the methylation level of the promoter region
[0096] First, genomic DNA was isolated using TiangenDP304 kit, and DNA degradation and contamination were monitored using agarose gel. DNA purity was measured with a spectrophotometer (IMPLEN, CA, USA). The DNA concentration was confirmed using the DNA Analysis Kit in the Qubit® 2.0 Flurometer, sonicated with a Covaris S220 to 200-300bp, followed by end repair and adenylation, and a total of 5.2 μg of genomic DNA fragments spiked with 26 ng λ DNA change. Ligate cytosine methylated barcodes to the sonicated DNA according to the kit manufacturer's instructions. The ligated DNA fragments were treated twice with bisulfite using the EZ DNA Methylation-Gold™ Kit (Zymo Research). Bisulfite-treated single-stranded DNA fragments were subjected to PCR amplification using the KAPA HiFi HotStartUracil + ReadyMix (2X) kit. Library concentration was quantified by Qubit 2.0 fluorometer (Life Technologies, CA, U...
Embodiment 2
[0100] Example 2 Obtaining the Methylation Score of the Promoter Region
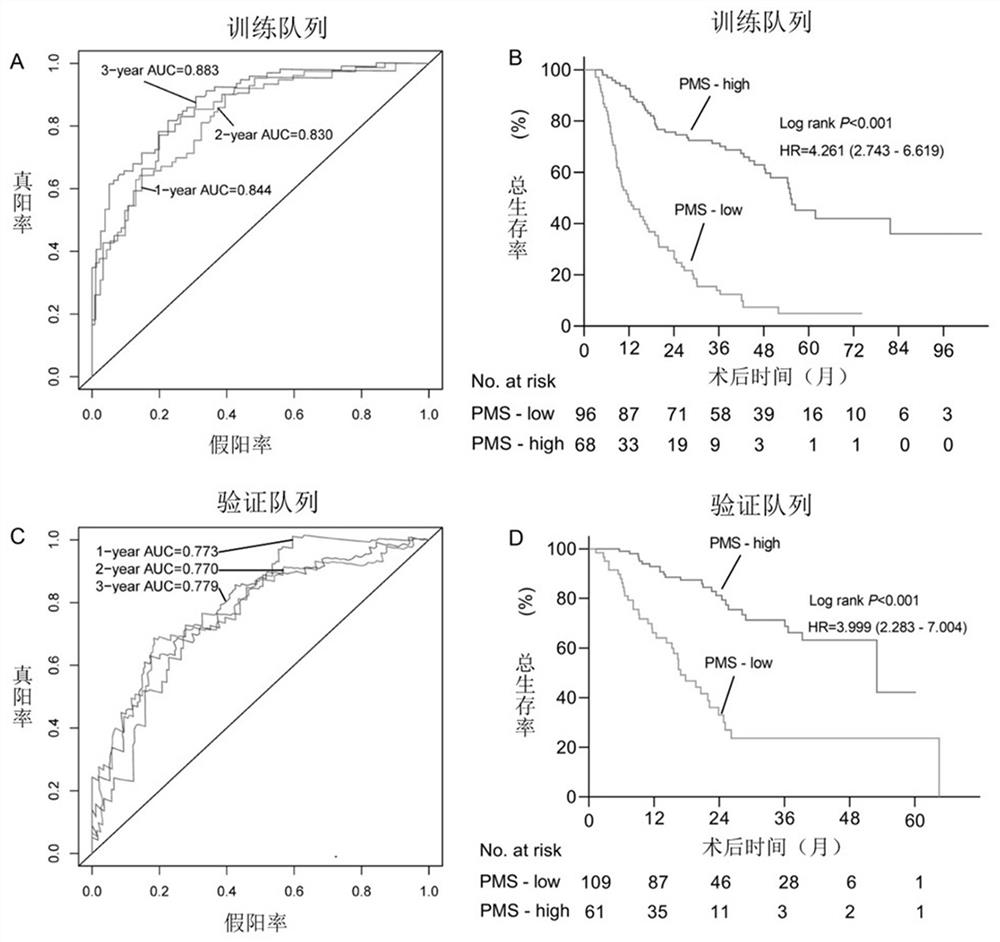
[0101] Using the 24 promoter regions shown in Table 1, calculate PMS (promoter methylation score, promoter region methylation score). PMS is the sum of the product of the methylation level of each candidate promoter region and the corresponding coefficient. The prognosis assessment model for patients with intrahepatic cholangiocarcinoma has one or more optimal thresholds, and the optimal thresholds can correspond to the scores calculated by the model for assessing the prognosis of patients with intrahepatic cholangiocarcinoma to one of high PMS and low PMS. The group is either a group of high PMS, medium PMS and low PMS.
[0102] The detailed formula of PMS is shown in formula (1).
[0103] With 164 patients in the training cohort, the C-index of Overall Survival (OS) was 0.772 (95% CI: 0.731-0.813), and the optimal threshold of PMS was -11.20716.
[0104] With 170 patients in the validation cohort, t...
experiment example
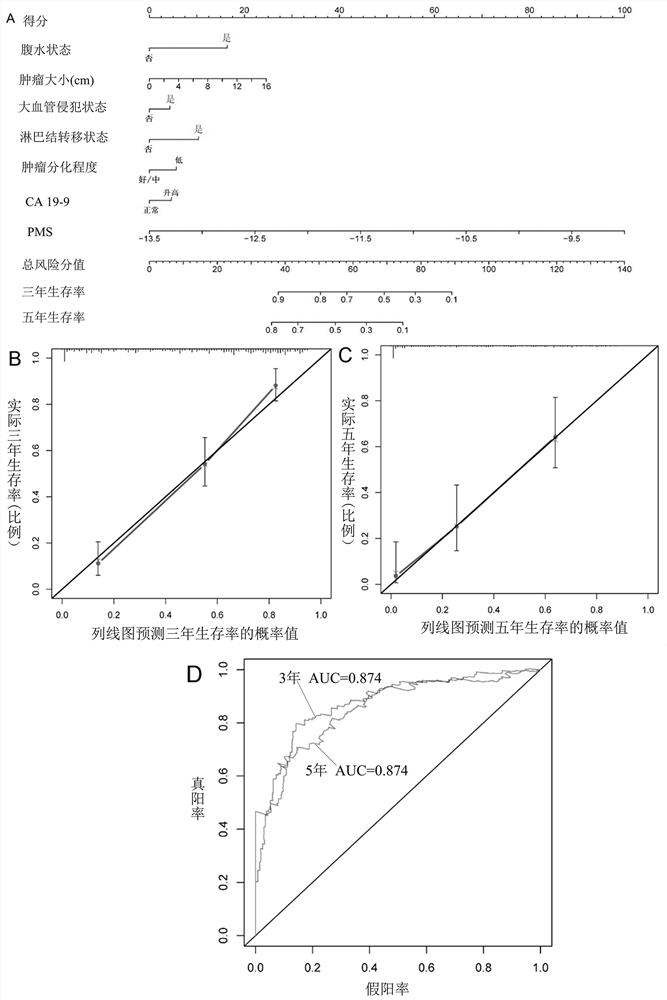
[0111] Experimental example: PMS predictor, WCHSU predictor, JHUSM predictor, EHBSH predictor, AJCC TNM predictor, WCHSU-PMS factor effectiveness comparison
[0112] PMS predictors are as described in Example 1 and Example 2.
[0113] WCHSU: ascites status, tumor size, microvascular invasion status, lymph node metastasis status, tumor differentiation degree and CA19-9 carbohydrate antigen concentration, respectively.
[0114] JHUSM: age, tumor size, tumor number, lymph node status, microvascular invasion status, and liver cirrhosis (Y Wang et al, Prognostic Nomogram for Intrahepatic Cholangiocarcinoma After Partial Hepatectomy, J. Clin. Oncol. 31(2013) 1188-1195).
[0115] EHBSH: See O Hyder, H Marques et al, A Nomogram to Predict Long-term Survival After Resection for Intrahepatic Cholangiocarcinoma, JAMA Surg. 149(2014)432.
[0116] AJCC TNM: T staging mainly includes three main factors: tumor number, vascular invasion, and direct extrahepatic invasion; N staging is based o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com