Methods and compositions for homology directed repair of double strand breaks in plant cell genomes
A double-strand break and homologous recombination technology, applied in the field of molecular biology, can solve problems such as low specificity, high preparation cost, and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0496] Example 1: Vector construction
[0497] Maize vector for SDN3
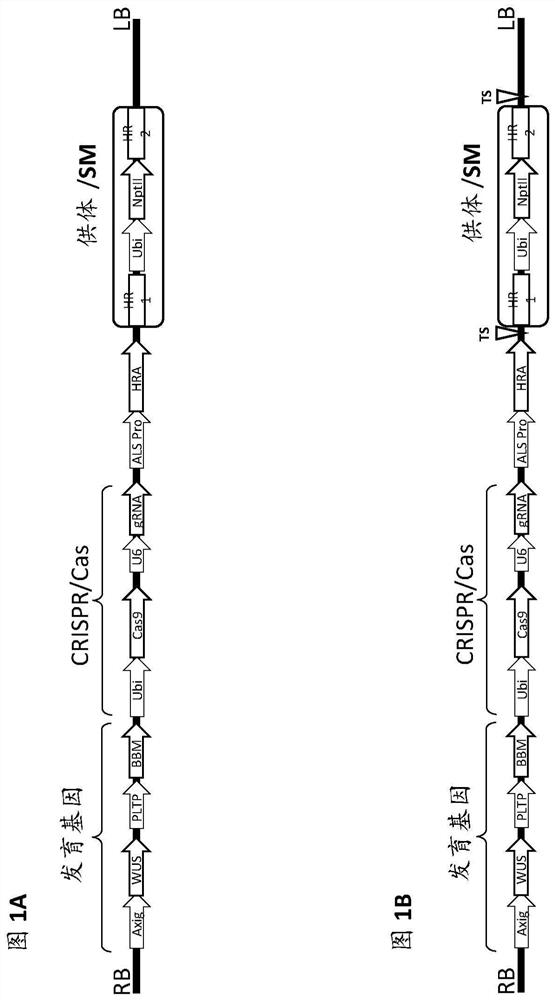
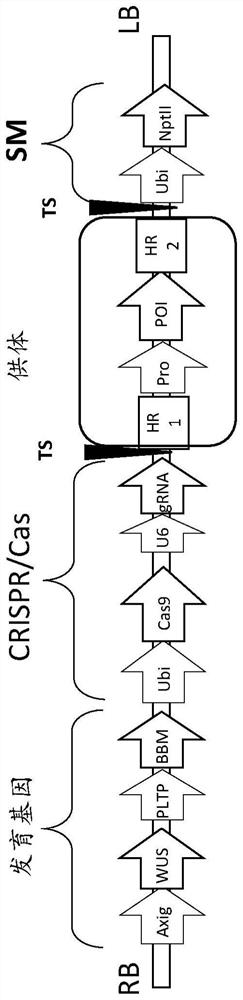
[0498] The T-DNA vector used in these experiments was Figure 5-8 Vectors 1-4 shown in and given as SEQ ID NOs: 46-49, respectively.
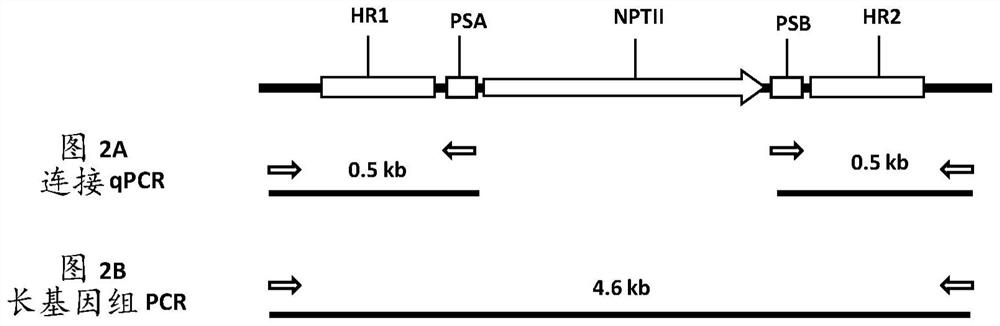
[0499]Each vector contains morphogenic factors (WUS and ODP2 under the Axig and PLTP promoters, respectively), cas9 driven by the ubiquitin promoter, and a target site operably linked to the maize U6 polymerase III promoter SEQ ID NO: 17 gRNA of dot (TS) sequence (SEQID NO:30), and two selectable marker genes - herbicide resistance ALS under native maize ALS and ubiquitin promoters (SEQID NO:20 and SEQID NO:8), respectively (HRA SEQ ID NO: 21) and neomycin phosphotransferase II (NptII, DNA SEQ ID NO: 24). The nptII gene driven by the ubiquitin promoter is flanked by a DNA fragment homologous to the sequence at the TS region (HR arms SEQ ID NO: 32 and SEQ ID NO: 33 for genotype A; HR arm SEQ ID NO for genotype B : 34 and SEQ ID NO: 35; for the HR arm of genotype C, SEQ ...
example 2
[0511] Example 2: Cell Transformation
[0512] Methods of Agrobacterium-mediated transformation of cells are known in the art (see, eg, US Patent Nos. 5,563,055 and 5,981,840). In one example, the method described in US 20170121722 A1 (published May 4, 2017) was used.
[0513] Prepare the Agrobacterium master plate.
[0514] Master plates were made by streaking Agrobacterium tumefaciens with binary donor vectors into solid 12V medium from -80°C frozen aliquots and incubating at 28°C for 2–3 days in the dark.
[0515] Grow Agrobacterium on solid media.
[0516] Pick single or multiple Agrobacterium colonies from the master plate and streak them onto a second plate containing 810I medium and incubate for 1-2 days at 28 °C in the dark. Add Agrobacterium infection medium (700 medium; 5 ml) and 100 mM 3′-5′-dimethoxy-4-hydroxyacetophenone (acetosyringone; 5 μL) to a 14 mL conical tube in a fume hood . About 3 full rings of Agrobacterium from the second plate were suspended...
example 3
[0529] Example 3: Plant regeneration
[0530] maize regeneration
[0531] Sixteen days later, embryos with healthy somatic embryos produced in Example 2 were transferred to regeneration medium.
[0532] In one example, the embryos were treated with Agrobacterium, and the selected embryos were transferred to 605T medium (not selected in the first week) one day later, with 0.1 mg / l ethametsulfuron-methyl containing AA in the 605T medium ( Early selection with AA) or in 605T medium with 0.1 mg / l ethametsulfuron (early selection without AA). For the next transfer, the selected embryos were moved to their respective maturation medium. For eventual transfer to rooting medium, single event selected seedlings were removed. For this experiment, the total time elapsed from Agrobacterium infection to the greenhouse was 48 days.
[0533] In another example, embryos were treated with Agrobacterium in liquid for 5 minutes and then co-cultured on 710I medium for one day. At this point...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com