Application of CRISPR (clustered regularly interspaced short palindromic repeats) gene editing technology in treatment of thalassemia
A base and methylation technology, applied in the field of gene editing, can solve the problems of low efficiency of plasmid delivery system, inability to achieve Cas9/sgRNA, and inability of lentivirus infection to achieve editing efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1 Design of sgRNA and Verification of Editing Efficiency
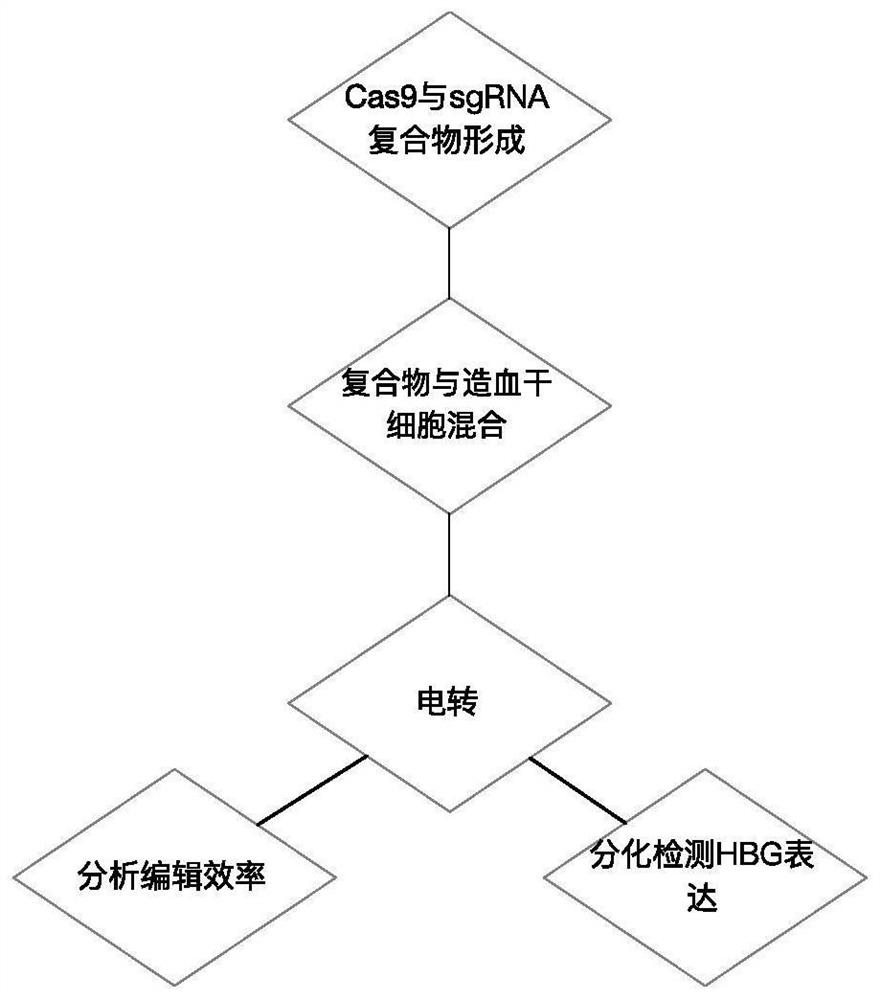
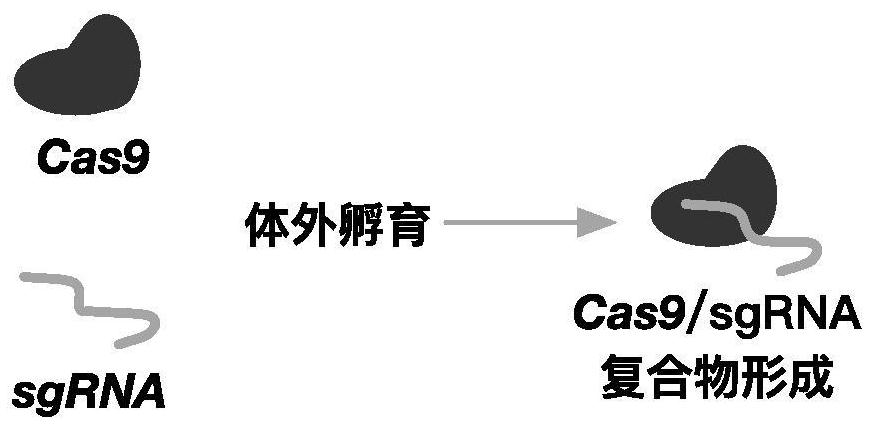
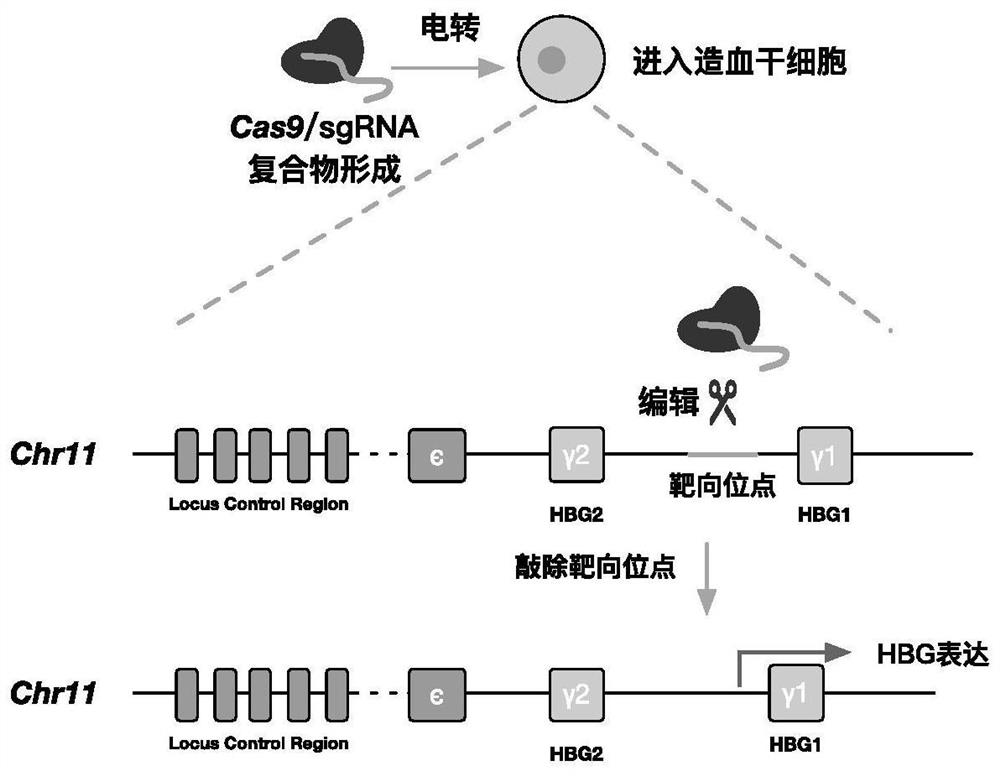
[0039] In this example, the complex of Cas9 and sgRNA was introduced into human hematopoietic stem cells by electroporation. The amino acid sequence of Cas9 is shown in SEQ ID No.5. The method flow is as follows figure 1 shown. Specifically, such as Figure 2-3 As shown, Cas9 and sgRNA can form a complex by in vitro incubation, and after electroporation into hematopoietic stem cells, they can target the upstream elements of the HBG1 gene for cleavage to up-regulate the expression of HBG.
[0040] In this embodiment, two sgRNAs are designed, sg-HBG1 and sg-HBG2, the targeting sequence of sg-HBG1 is CTTGTCAAGGCTATTGGTCA, and the targeting sequence of sg-HBG2 is CTTGACCAATAGCCTTGACA. Two chemically modified and unmodified sgRNAs were synthesized in vitro. Here, the chemical modification refers to the 1st, 1-2 or 1-3 base at the 5' end of the sgRNA targeting sequence, and / or the 1st, 1-2 or 3' base at th...
Embodiment 2
[0043] Example 2 Editing of the HBG promoter site in human hematopoietic stem cells
[0044] In this embodiment, a Cas9 and sgRNA complex is used to introduce the complex in human hematopoietic stem cells by electroporation, the amino acid sequence of the Cas9 is shown in SEQ ID No.5; the sgRNA sequence includes a targeting sequence and a backbone 序列,所述靶向序列为5'CTTGTCAAGGCTATTGGTCA3'(SEQ ID No.1),所述骨架序列为5'GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGC3'(SEQ ID No.2);所述sgRNA序列为5'CTTGTCAAGGCTATTGGTCAGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGC3'(SEQ ID No .3).
[0045] In this embodiment, the 1-3 bases at the 5' end and the 1-3 bases at the 3' end of the sgRNA (SEQ ID No.3) are methylated; in other embodiments, It is also possible to carry out methylation modification or any other chemical modification at the 1-5th base at the 5' end of the sgRNA or at any one or any number of bases at the 1-5th base at the 3' end, such...
Embodiment 3
[0050] Example 3 Detection of HBG expression after gene editing
[0051] The cells of the experimental group and the control group were cultured in differentiation medium 1 (stemspan containing: 10% FBS, IL-3: 10ng / ml, SCF: 50ng / ml, EPO: 1U / ml) for 7 days, and then centrifuged at 300g for 10 minutes Seed to pellet cells, change into differentiation medium 2 (stemspan containing 30% FBS, EPO: 3U / ml) and culture for 2 days. Cells were centrifuged at 300g. Total mRNA was extracted and reverse-transcribed to obtain cDNA.
[0052] Use HBG gene forward primer 5'GATGCCATAAAGCACCTGGATG3', reverse primer 5'TTGCAGAATAAAGCCTATCCTTGA3' for QPCR amplification of HBG mRNA, use HBB gene forward primer 5'TCAAGGGCACCTTTGCCACAC3' reverse primer 5'TGATAGGCAGCCTGCACTG3' for QPCR detection of HBG mRNA relative to The expression level of HBB gene. Such as Figure 7 As shown, after editing, the content of HBG mRNA relative to HBB was significantly increased in the experimental group.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com