Specific primer and test kit for detecting pathogenic variation of deafness gene based on multiplex PCR and high-throughput sequencing technology and application
A technology of technical detection and deafness gene, which is applied in the field of genetic detection, can solve problems such as time-consuming and laborious, high technical requirements, and restrictions on wide application, and achieve the effects of reducing costs, simplifying operation steps, and improving detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
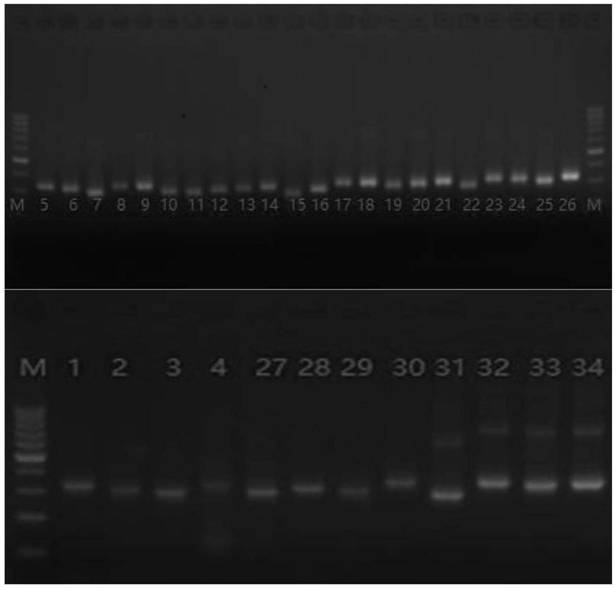
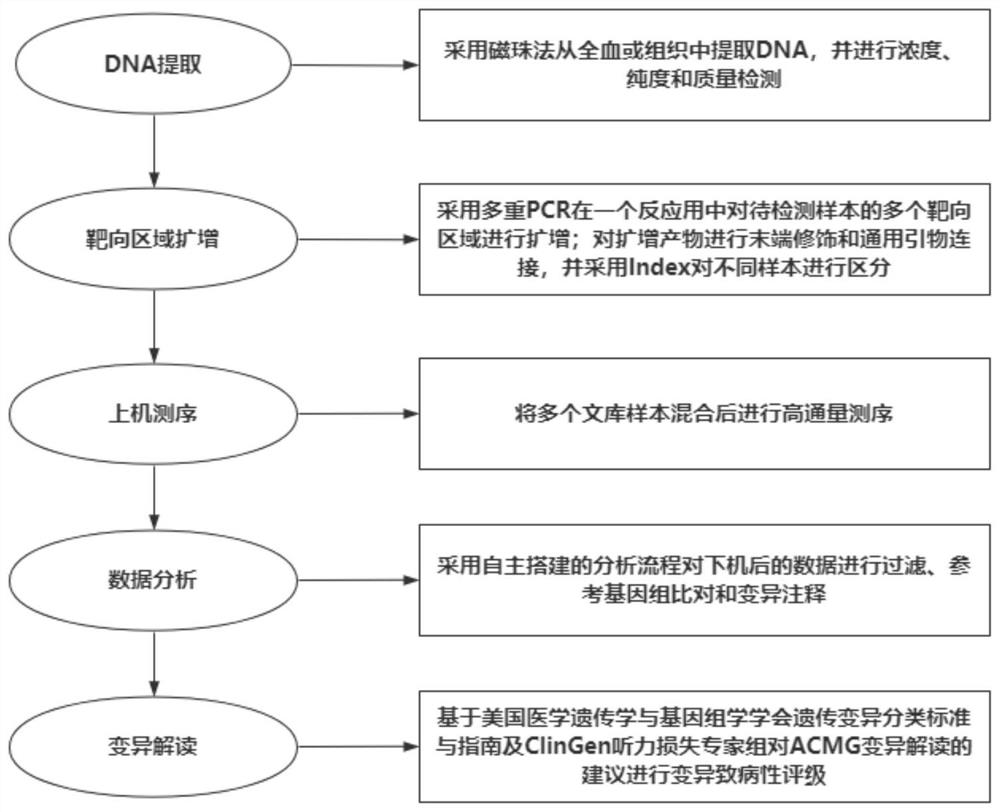
Examples
Embodiment 1
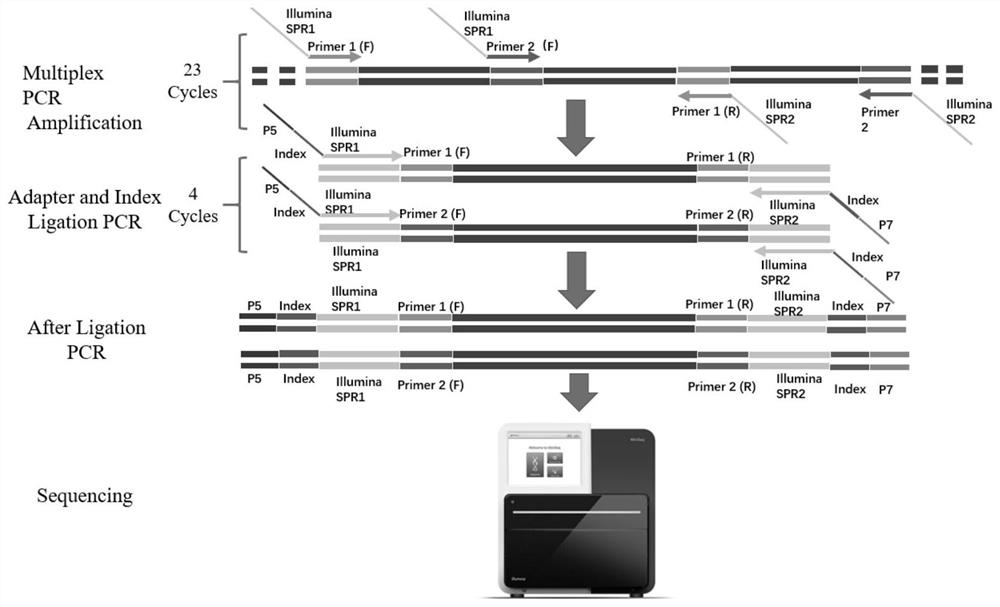
[0030] Example 1 uses specific primers to amplify the target region:
[0031] Since the primer amplification regions are too close or overlap each other, they need to be divided into two primer pools for PCR respectively. The primers are library amplification primer I (18 pairs of primers) and library amplification primer II (16 pairs of primers).
[0032] Thaw deafness-specific primers I, deafness-specific primers II, multiplex PCR enzyme mixture, positive quality control (PC), negative quality control (NC) and blank quality control (BC) and mix them upside down ten times after thawing , centrifuge briefly, and store on ice for later use.
[0033] 1) DNA template preparation: quantify the extracted nucleic acid to 1-10ng / μl, and the final volume is greater than 11μl;
[0034] 2) Take the deafness-specific primer I and multiple PCR enzyme mixture in a PCR tube to prepare the following reaction (denoted as: library amplification product 1):
[0035] Note: Take 5.5 μl each of th...
Embodiment 2
[0044] Example 2 Amplified product end repair, universal primer and Index connection
[0045] 1) In this step, the adapter will be connected to the end of the PCR product. After thawing the amplification enzyme mixture, the universal end adapter and the required index adapter, invert it up and down ten times to mix well, centrifuge briefly, and put it on ice for later use;
[0046] 2) Prepare the following reaction in a new PCR tube (marked as: PCR Mix tube):
[0047]
[0048] 3) Use a pipette to blow and mix 10 times, and place in a palm centrifuge for 5 seconds;
[0049] 4) Transfer the two tubes of reaction solutions of library amplification product 1 and library amplification product 2 to PCR Mix tubes in sequence, mix well by blowing and blowing for 10 times with a pipette, and centrifuge in a palm centrifuge for 5 seconds;
[0050] 5) Put the PCR tube in the PCR instrument and perform the following reaction:
[0051]
[0052]
[0053] 6) After the amplificatio...
Embodiment 3
[0054] Example 3 Purification
[0055] Purify the reaction product using the recommended purification beads Vazyme DNA Clean Beads:
[0056] 1) After the magnetic beads are equilibrated to room temperature, vortex to mix the DNA Clean Beads;
[0057] 2) According to the ratio of 1:0.8, pipette 64μl DNA Clean Beads into 80μl PCR product, vortex or use a pipette to gently pipette 10 times to mix thoroughly;
[0058] 3) Incubate at room temperature for 5 minutes;
[0059] 4) Centrifuge the PCR tube in a palm centrifuge for 5 seconds and place it in a magnetic stand to separate the magnetic beads and liquid. After the solution is clarified (about 5 minutes), carefully remove the supernatant;
[0060] 5) Keep the PCR tube always placed in the magnetic stand, add 180 μl of freshly prepared 80% ethanol to rinse the magnetic beads, incubate at room temperature for 30 seconds, and carefully remove the supernatant;
[0061] 6) Repeat the above step 5), rinse twice in total;
[0062]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com