A sgRNA targeted to knock out the sst gene and its CRISPR/Cas9 system and application
A genetic and targeting technology, applied in DNA/RNA fragments, applications, genetic engineering, etc., can solve problems such as low efficiency, cumbersome steps, and long time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Construction of Cas9 sgRNA knockout plasmid
[0048] 1.1 Screening and synthesis of sgRNA
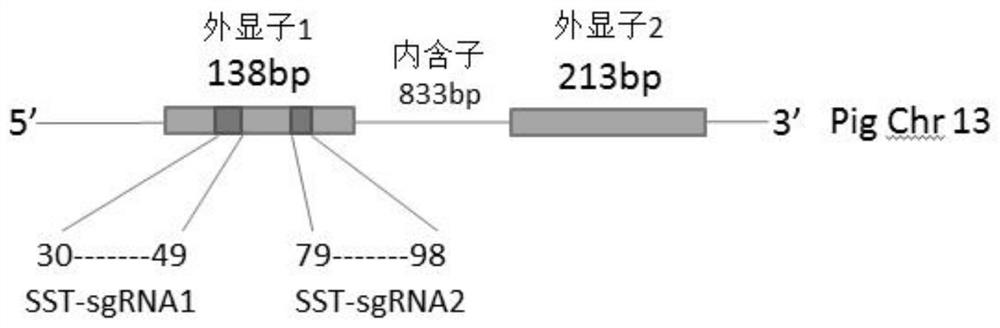
[0049] Select the first exon of the porcine SST gene (GenBank accession number: NC_010455.5) as the targeting region, and use the website (http: / / crispor.tefor.net) to design sgRNA for the targeting region. The design conditions are: ① The length of the sgRNA is 20nt, the core sequence is NNNNNNNNNNNNNNNNNNNN(20)-NGG (SEQ ID No.6); ② Add CACCG to the 5' end of the sense strand template, and add AAAC to the 5' end of the antisense strand template to make the oligonucleotide chain and the vector plasmid The cohesive ends are complementary. Finally, two suitable targets were selected and named as SST-sgRNA1 and SST-sgRNA2. CACC was added to the 5' end of the coding strand template, and AAAC was added to the 3' end of the non-coding strand template to complement the cohesive ends formed after digestion with BbsI, and two pairs of CRISPR oligonucleotide chains were designed.
[005...
Embodiment 2
[0053] vector construction
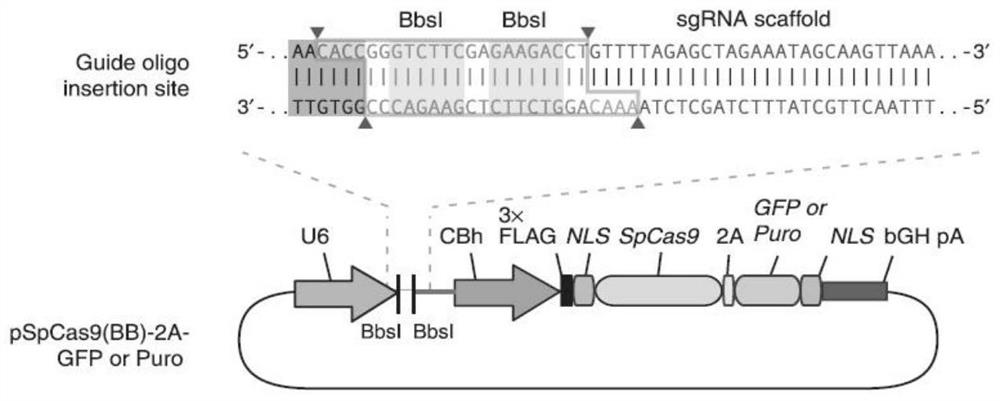
[0054] 1.2.1 Use BbsI to digest the pSpCas9(BB)-2A-Puro plasmid, and the digestion reaction system is as follows:
[0055] Element Amount added pSpCas9(BB)-2A-Puro plasmid 1μg BbsI enzyme 2μL FDbuffer 2μL wxya 2 o
to 50μL Total 50μL
[0056] Operated on ice, the above components were added sequentially, after thorough mixing, enzyme digestion was carried out in a circulating water bath at 37°C for 2 hours.
[0057] 1.2.2 Recover the above digested products according to the instructions of Novizan Gel Recovery Kit.
[0058] 1.2.3 sgRNA oligo annealing and double strand formation
[0059] Dilute each pair of oligonucleotides synthesized by the company to 10 μmol / L and mix according to the following ratio: 16 μL ddH 2 O, 2 μL 10×NEB Buffer 3, 1 μL sense strand, 1 μL antisense strand, denature at 95°C for 5 minutes after mixing, and then naturally anneal at room temperature for 1 hour to form do...
Embodiment 3
[0065] Transfection of porcine kidney fibroblasts by electroporation
[0066] 1.1 Main reagents and materials
[0067] Male Large White piglets aged 1-3 days were from the experimental pig farm of the Institute of Animal Husbandry and Veterinary Medicine, Hubei Academy of Agricultural Sciences; G418 antibiotics were purchased from Sigma; DMEM, DPBS, fetal bovine serum, DMSO and other cell culture and cryopreservation reagents were purchased from Gibco. Electrotransfer buffer was prepared by the laboratory itself.
[0068] 1.2 Isolation and culture of porcine kidney fibroblasts
[0069] Take a newborn 1-3d male large white pig, deeply anesthetized by intraperitoneal injection of pentobarbital sodium (50 mg); fully clean and disinfect the large white pig body surface with 75% alcohol, and then use sterilized medical surgical instruments to remove the double The side kidneys were soaked in 75% alcohol and moved to a clean bench.
[0070] The kidney was washed repeatedly with D...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com