Uridine phosphatase mutant and application thereof
A technology of uridine phosphatase and mutant, applied in the fields of genetic engineering and enzyme catalysis, can solve the problem of low activity of nicotinamide substrate, and achieve the effect of improving enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1 Construction of wild-type uridine phosphatase gene recombinant Escherichia coli
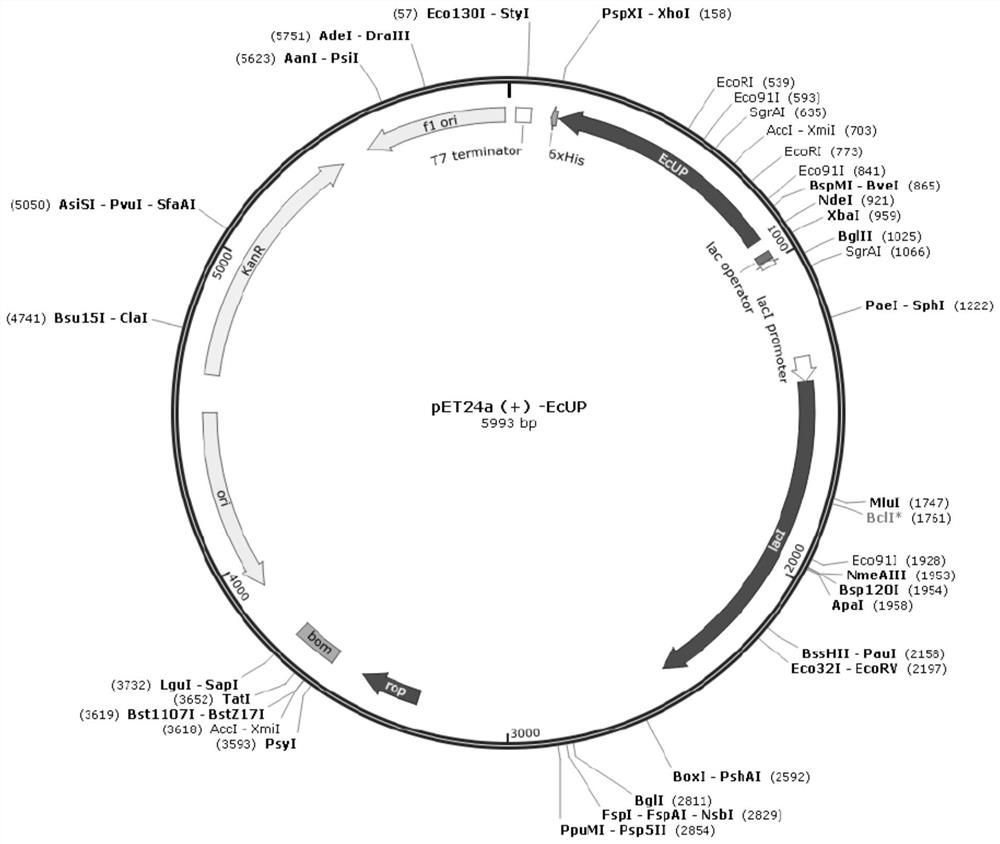
[0048] For the wild-type uridine phosphatase derived from Escherichia coli K-12substr.MG1655 (the published amino acid GenBank sequence number is P12758), that is, SEQ ID NO: 1, codon optimization is carried out on this basis, and the whole gene synthesis coding gene sequence SEQ ID NO: 2, and design restriction endonuclease sites Nde I and XhoI at both ends of the gene, subclone into the corresponding sites of the vector pET24a (Novagen), obtain the recombinant plasmid pET24a-EcUP, see the plasmid map figure 1 . The recombinant plasmid pET24a-EcUP was transformed into the expression host Escherichia coli BL21(DE3) to obtain the recombinant Escherichia coli pET24a-EcUP / BL21(DE3) expressing wild-type uridine phosphatase SEQ ID NO:1.
Embodiment 2
[0049] Example 2 Construction of Nicotinamide Ribokinase Recombinant Escherichia coli
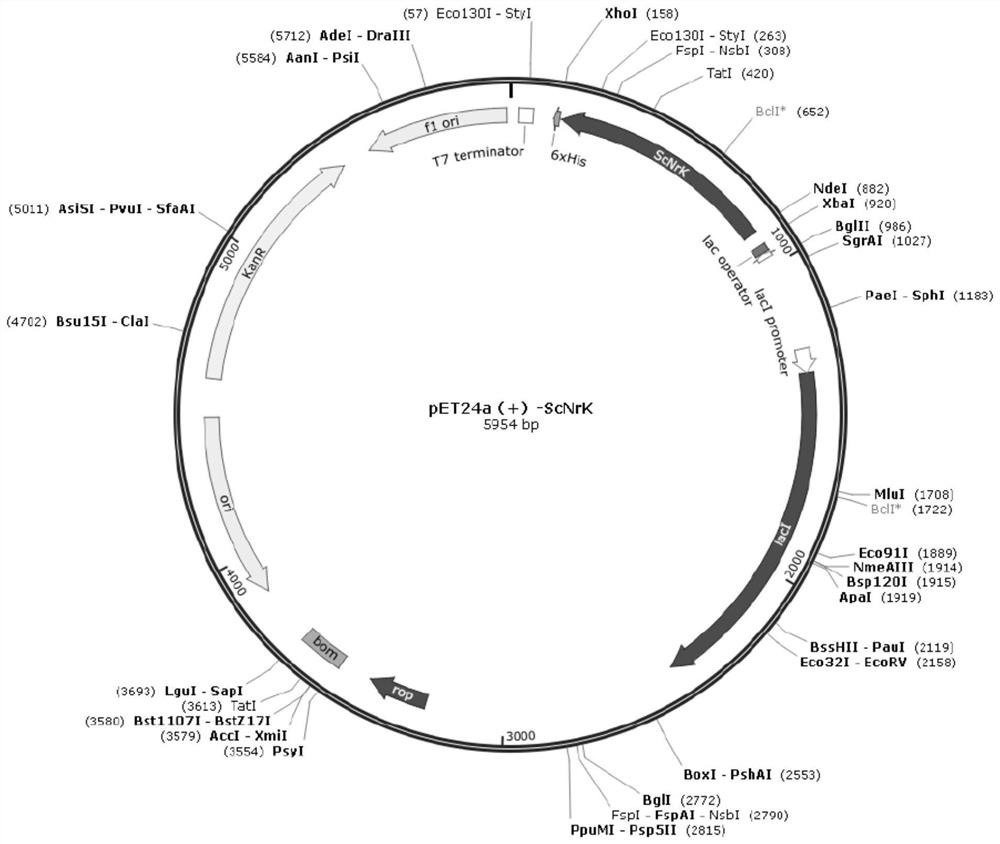
[0050] For the nicotinamide ribokinase coding gene derived from Saccharomyces cerevisiae S288C (the published nucleic acid GenBank sequence number is NM_001182967.1), that is, SEQ ID NO: 5, the whole gene was synthesized, and restriction endonuclease sites were designed at both ends of the gene Point Nde I and XhoI, subclone into the corresponding site of the vector pET24a (Novagen), obtain the recombinant plasmid pET24a-ScNrK, see the plasmid map figure 2 . The recombinant plasmid pET24a-ScNrK was transformed into the expression host Escherichia coli BL21(DE3) to obtain the recombinant Escherichia coli pET24a-ScNrK / BL21(DE3) expressing nicotinamide ribokinase.
Embodiment 3
[0051] Example 3 Construction of uridine phosphatase mutant recombinant Escherichia coli
[0052] The wild-type uridine phosphatase was mutated by site-directed combinatorial mutation.
[0053] The pET24a-EcUP plasmid was used as a template, and 162F / 195R and 195F / 220&221R were used as primer pairs for PCR amplification. The primers were designed as follows:
[0054] 162F: 5'-GTGTTACCGCTTCTTCTGACACCGGTTACCCGGGTCAGGAAC-3',
[0055] 195F: 5'-GCAGGCTATGGGTGTTATGAACGCAGAAATGGAATCTGCTACCCTG-3',
[0056] 195R: 5'-CAGGGTAGCAGATTCCATTTCTGCGTTCATAACACCCATAGCCTGC-3',
[0057] 220&221R:
[0058] 5'-GATTTCCTGCTGGGTACGGTTAGAACCAACACCAGCAACCATACCA-3'.
[0059] The PCR reaction system includes: 50pmol each primer, 10ng plasmid template, 1×KOD neo plus buffer, 0.2mM dNTP, 1.5mM MgSO 4 , KOD neo plus 1U, add ddH 2 O to 50 μL of the total system.
[0060] The PCR amplification conditions were: 95°C for 5min; 98°C for 15s, 57°C for 30s, 68°C for 30s / kbp, 30 cycles; 68°C for 10min.
[006...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com