Nucleic-acid visual detection kit for novel coronavirus SARS-CoV-2
A coronavirus, visual detection technology, applied in the direction of DNA / RNA fragments, recombinant DNA technology, microbial measurement / inspection, etc., can solve the problems of high technical requirements, unfavorable clinical application, expensive instruments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Preparation of Visual Detection Kit for Novel Coronavirus SARS-CoV-2 Nucleic Acid
[0034] (1) RT-LAMP primer design, synthesis and preparation of primer set solution:
[0035] Log in to the novel coronavirus gene sequence database of the National Genome Science Data Center (https: / / bigd.big.ac.cn / ncov). Search and download novel coronavirus, other coronaviruses and other pneumonia-causing viruses (influenza virus, respiratory syncytial virus, rhinovirus, adenovirus, parainfluenza virus, enterovirus, herpes simplex virus, human metapneumovirus, reovirus , measles virus, cytomegalovirus, etc.) genome sequences, using CLUSTAL, DNASTAR, BIOEDIT and other bioinformatics software to compare and analyze the genome sequences. The analysis found that there was no significant variation in the genome sequences of the recently popular new coronavirus SARS-CoV-2 isolates, so the representative strain of the new coronavirus SARS-CoV-2, BetaCoV / Wuhan / WH-01 / 2019, was selec...
Embodiment 2
[0051] Example 2: Optimization of the detection temperature of the new coronavirus SARS-CoV-2 nucleic acid visual detection kit
[0052] To each RT-LAMP reaction system (25ul), add 12.5μL of 2×LAMP reaction solution, 7ul of primer set aqueous solution, 1ul of positive template, and make up the volume to 25ul with sterile DEPC water. The final concentration of each component in each 25ul RT-LAMP reaction system is 20mM Tris-HCl (pH8.8), 10mM KCl, 8mM MgSO 4 , 10mM (NH 4 ) 2 SO 4 , 0.1% Tween 20, 0.8M betaine, 1.4mM dNTPs, 8U Bst DNA polymerase, 200U AMV reverse transcriptase, 2.5U RNase inhibitor, 0.25mMHNB, the moles of each primer are: primers F3, B3 each 5pmol, primers FIP, BIP each 40pmol. A negative control and a blank control were set up in the experiment.
[0053] The temperature of RT-LAMP reaction was set at 58, 59, 60, 61, 62, 63, 64, and 65 °C, respectively, and the reaction time was 50 minutes. Then, the reaction was terminated at 85 °C for 5 minutes. Visually...
Embodiment 3
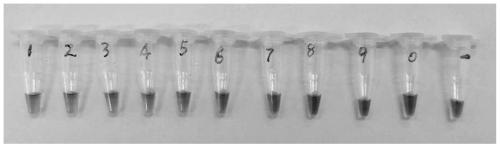
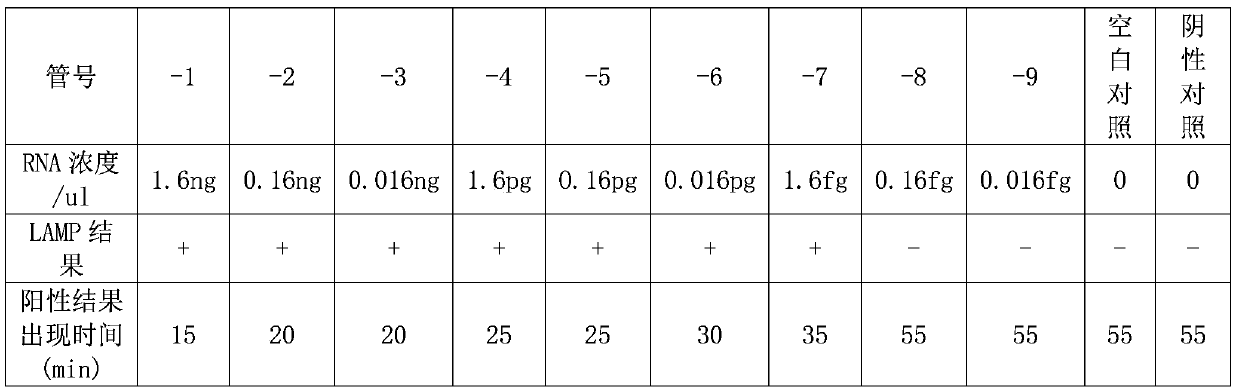
[0057] Example 3: Detection time optimization and sensitivity determination of SARS-CoV-2 nucleic acid visual detection kit
[0058] Positive templates were diluted 10-fold with DEPC water. The amount of positive template in tubes 1# to 9# was 1.6ng, 0.16ng, 0.016ng, 1.6pg, 0.16pg, 0.016pg, 1.6fg, 0.16fg, 0.016fg / ul, respectively. Tubes 10# and 11# are negative control and blank control, respectively.
[0059] The RT-LAMP reaction system is the same as the reaction system in Example 2. That is, to each RT-LAMP reaction system (25ul), add 12.5μL of 2×LAMP reaction solution, 7ul of primer set aqueous solution, 1ul of positive templates of different concentration gradients, and make up the volume to 25ul with sterilized DEPC water. The final concentration of each component in each 25ul RT-LAMP reaction system is 20mM Tris-HCl (pH8.8), 10mM KCl, 8mM MgSO 4, 10mM (NH 4 ) 2 SO 4 , 0.1% Tween 20, 0.8M betaine, 1.4mM dNTPs, 8U Bst DNA polymerase, 200U AMV reverse transcriptase, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com