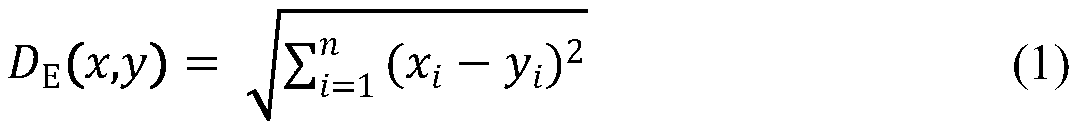Construction method of medium-temperature anaerobic digestion removal rate prediction model for PPCPs organic chemicals
A technology of organic chemicals and anaerobic digestion, which is applied in the field of construction of prediction model of removal rate of thermoanaerobic digestion, can solve the problem that removal efficiency has not been evaluated, and achieve the effect of saving manpower, avoiding actual measurement and research, and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1 Construction of prediction model for removal rate model of mesothermal anaerobic digestion of PPCPs organic chemicals
[0036] (1) Data collection, setting training set and verification set sample compounds;
[0037] The data on the removal rate of 44 organic chemicals in sludge under the condition of mesophilic anaerobic digestion were queried from the literature (sludge residence time SRT = 25d, PH = 8.0; from the literature Phase distribution and removal of pharmaceuticals and personal care products during anaerobic sludge digestion . Journal of Hazardous Materials, 2013, 260, 305–312). A: removal rate ≥ 60% can be removed; I removal rate < 60% can not be removed. The training set selected 35 sample compounds, and the verification set selected 9 sample compounds. The training set samples should be as diverse in structure as possible, and the activity coverage should be as large as possible, so that the model can have a wide range of applications and stron...
Embodiment 2
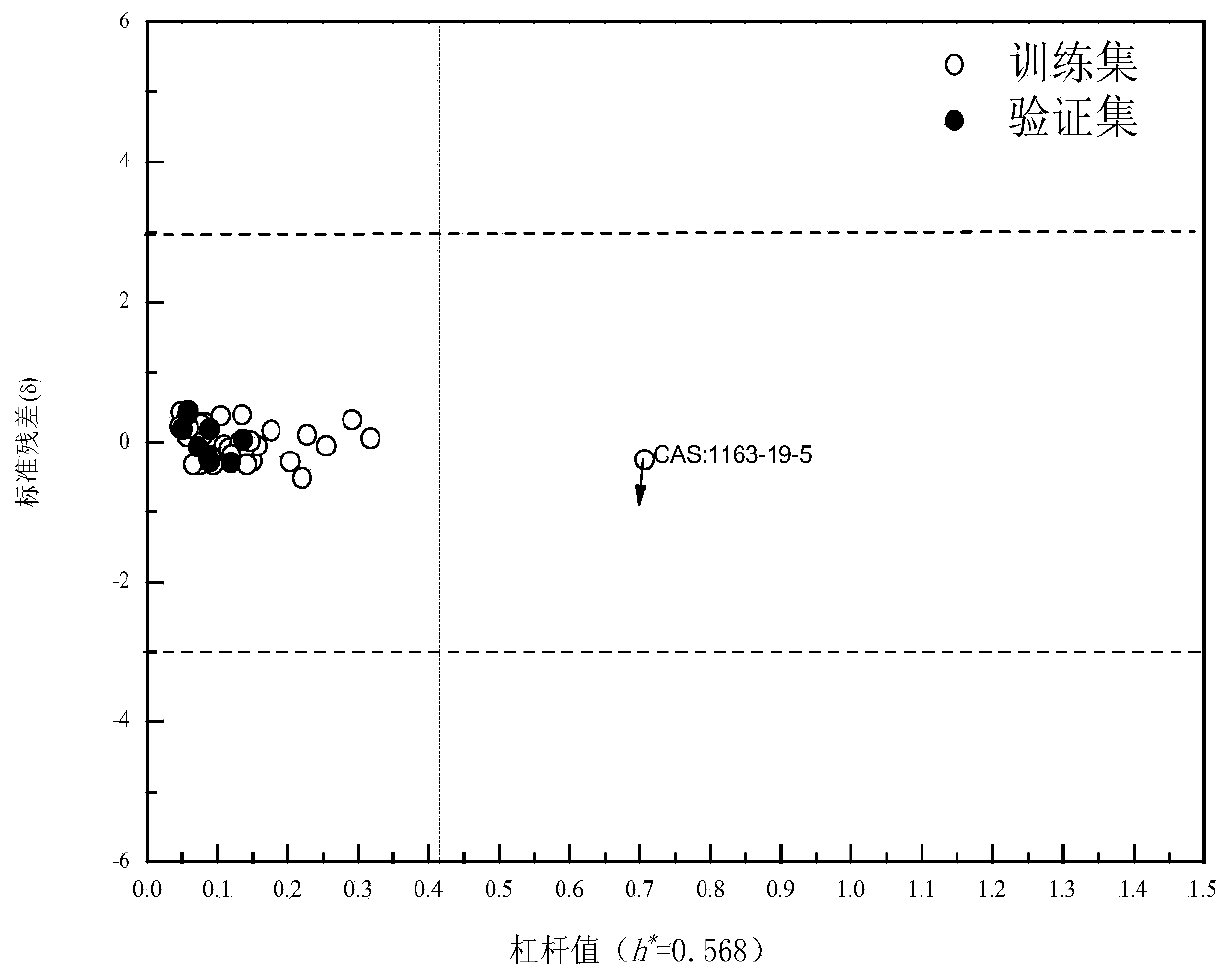
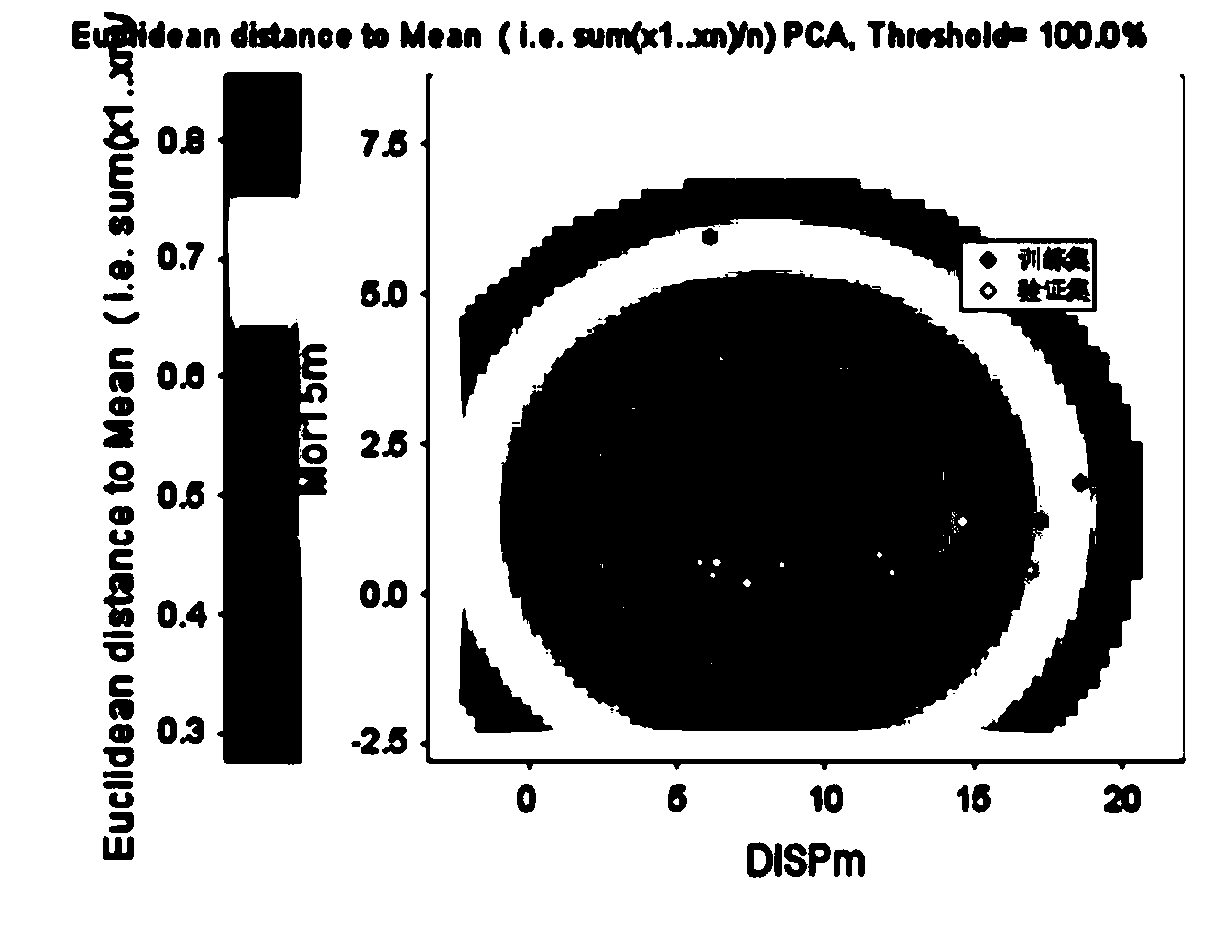
[0057] Example 2 Perform model application domain characterization on the constructed model
[0058] The domain of application of the model is defined using a Euclidean distance method and a leverage-based Williams diagram. Euclidean distance was calculated using AMBIT Discovery v0.04 software (http: / / ambit.sourceforge.net / download_ambitdiscovery.html). The Euclidean distance is calculated by the following formula:
[0059]
[0060] where μ is the mean value of the descriptor x.
[0061] The Williams diagram is composed of standard residuals (δ) and leverage values (in h i Indicates that i represents a different compound) defines a model application domain. δ is calculated using the following formula:
[0062]
[0063] The leverage value of the training set compound (leverage, h i ) can be obtained by the following formula:
[0064] h i =x i T (X T X) –1 x i (8)
[0065] where x i is a row vector of molecular structure descriptors for the i-th compound. ...
Embodiment 4
[0076] The mesophilic anaerobic digestion removal rate prediction of embodiment 4 azithromycin
[0077] Give azithromycin (SMILES: CN([C@H](C)[C@@H](O)[C@@](O)(C)[C@@H](CC)O1)C[C@ H](C)C[C@](O)(C)[C@H](O[C@H]3[C@H](O)[C@@H](N(C)C) C[C@@H](C)O3)[C@@H](C)[C@H](O[C@H]2C[C@@](C)(OC)[C@@H ](O)[C@H](C)O2)[C@@H](C)C1=O) to predict the removal rate of mid-temperature anaerobic digestion. First, according to the molecular structure of the chemical substance, four descriptors DI SPm, Mor15m, HATSe, O-060 were calculated using Dragon software; they were 3.274, 2.213, 8.685 and 0 respectively; Euclidean distance was calculated with each compound in the training set, and the calculated The three nearest training set compounds were clarithromycin, roxithromycin and trimethoprim. Hat is 0.311, and Euclidean distance is 0.369. Within the scope of the model application domain, this model can be used to predict the anaerobic digestion removal rate of ciprofloxacin:
[0078] The predicted re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com