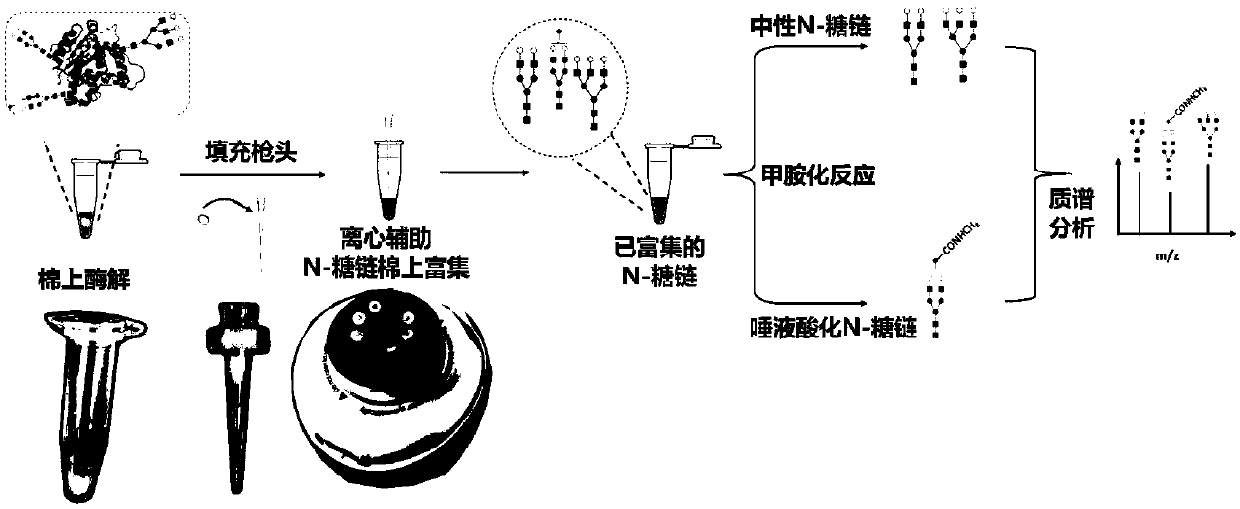
Method for rapid enzymolysis release, solid phase concentration and mass spectrometry of N-glycan
A sugar chain and enzymatic hydrolysis technology, which is applied in the field of glycoproteomics and glycomics analysis, can solve the problems of high price, long operation time, and low proportion of glycosylation modification, so as to improve the selectivity and binding speed of mass spectrometry analysis The effect of fast and strong affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
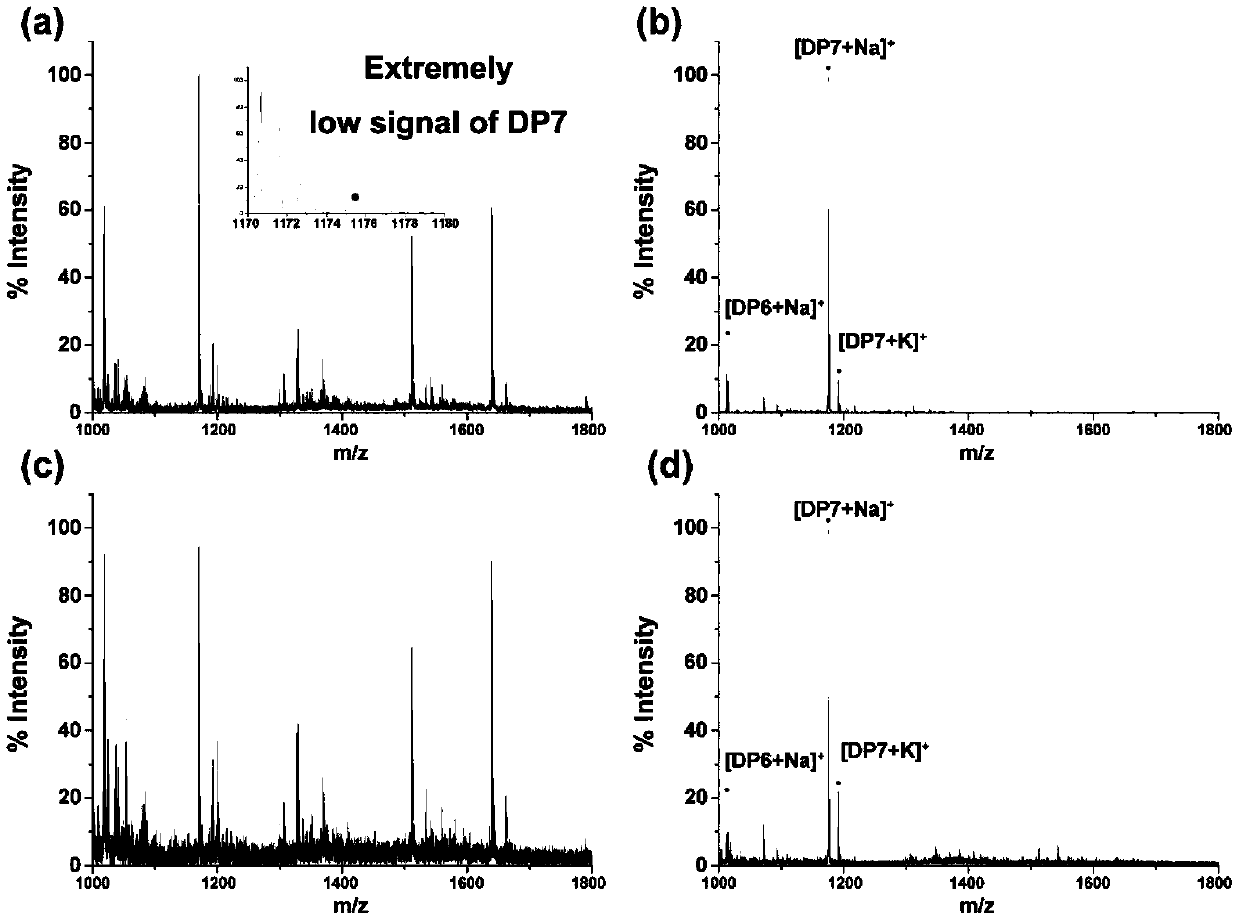
[0039] For solid-phase enrichment of samples containing free N-glycans
[0040] Weigh 3-7mg absorbent cotton material, fill it into the tip of the pipette, and place it on the centrifuge tube. Use 50-80 μL of water to activate once, centrifuge to remove the solution, the interval between each addition of liquid and centrifugation is 0.5-1 minute, the centrifugal force is 1500g, and the centrifugation time is 10-30 seconds, the same below; then use 50-80 μL 80 -85% acetonitrile aqueous solution was washed once, and the solution was removed by centrifugation. After the desalted N-glycan sample (DP7:BSA peptide = 1:1000) was lyophilized, it was redissolved with 80-160 μL of acetonitrile / trifluoroacetic acid / water solution, so that the volume fraction of acetonitrile in the solution after the reaction was 80-85 %, the volume fraction of TFA is 0-0.1%, the sample is loaded, and the solution is removed by centrifugation. Use 50-100 μL 75-85% acetonitrile / water solution to wash 4-6...
Embodiment 2
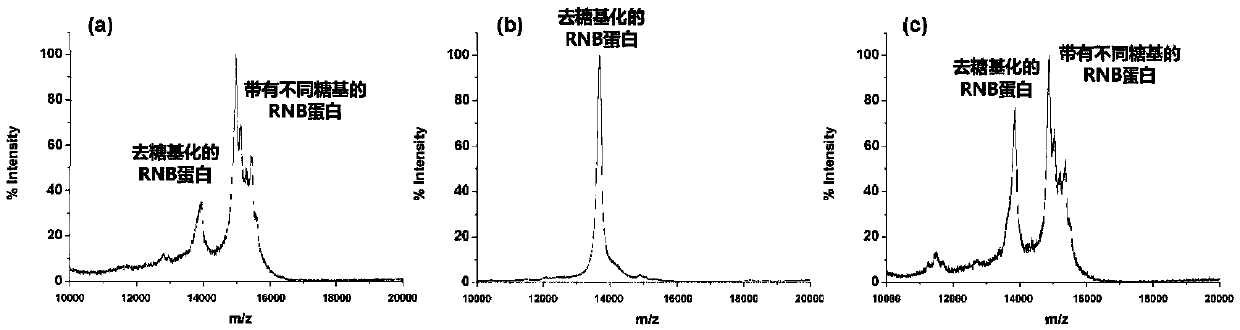
[0042] High-speed enzymatic digestion and solid-phase enrichment on cotton for N-glycoprotein-containing samples
[0043] Weigh 3-7 mg of absorbent cotton material and place it at the bottom of the centrifuge tube. After freeze-drying the N-glycoprotein sample to be processed (ASF:BSA enzymatic peptide = 1:50), redissolve it with 20-40 μL 25mM ammonium bicarbonate buffer to make the final concentration 10ng / μL-1000ng / μL , transferred to a centrifuge tube containing absorbent cotton material. Treat in a water bath at 100 degrees Celsius for 5 minutes, add 0.5 μL PNGase F enzyme after cooling, and enzymatically hydrolyze at 45-55 degrees Celsius for 0.5-1 hour. A certain volume of acetonitrile / trifluoroacetic acid / water solution is added to the solution after enzymolysis, so that the volume fraction of acetonitrile in the solution is 80-85%, the volume fraction of TFA is 0-0.1%, and the volume is 80-160 μL. Fill the pipette tip with absorbent cotton material and place it on th...
Embodiment 3
[0045] High-speed enzymatic digestion and solid-phase enrichment on cotton for complex samples of human serum
[0046] Weigh 3-7 mg of absorbent cotton material and place it at the bottom of the centrifuge tube. 1 μL of serum to be treated was diluted with 20-40 μL of 25 mM ammonium bicarbonate buffer, and then transferred to a centrifuge tube containing absorbent cotton material. Treat in a water bath at 100 degrees Celsius for 5 minutes, add 0.5 μL PNGase F enzyme after cooling, and enzymatically hydrolyze at 45-55 degrees Celsius for 0.5-1 hour. A certain volume of acetonitrile / trifluoroacetic acid / water solution is added to the solution after enzymolysis, so that the volume fraction of acetonitrile in the solution is 80-85%, the volume fraction of TFA is 0-0.1%, and the volume is 80-160 μL. Fill the pipette tip with absorbent cotton material and place it on the centrifuge tube. Load the remaining solution, centrifuge to remove the solution, the interval between each addi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com