Method for high-throughput rapid evaluation and/or screening of saline-alkali tolerant plants
A plant and salt-tolerant technology, applied in the direction of measuring devices, fluorescence/phosphorescence, color/spectral characteristics measurement, etc., can solve the problems of limited space and site area, long salt-tolerant occupation, long growth cycle, etc., and achieve simple equipment Easy to obtain, high energy and short experimental period
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Elaeagnus sativa with high salt tolerance was used as the experimental material, among which, Ale sativa (ALE) and Yinchuan (YC) were two provenances, and the salt tolerance of ALE provenance was slightly stronger than that of YC provenance.
[0028] NaCl solution configuration: 150mM NaCl solution is used for salt treatment, and a sufficient amount of salt solution needs to be prepared in advance according to the types of test tree species and the test repetition settings for future use.
[0029] Use branch scissors to cut out branches of the same part of Elaeagnus erae to be measured at the same age or growth time from the field, with a length of about 15-30 cm, soak in clean water until the experimental treatment (to prevent water loss), and use as much as possible , to ensure that the branches and leaves do not wilt while soaking in the salt solution.
[0030] The prepared plant branch to be tested is put into the 50ml centrifuge tube that fills 150mM NaCl solution ...
Embodiment 2
[0038] The test material is the bamboo willow whose salt tolerance is lower than that of Elaeagnus sativa, and the others are the same as in Example 1.
[0039] See Table 1 for the results of the total chlorophyll content and Fv / Fm of the bamboo willow blank control group and the salt treatment group at each treatment time point.
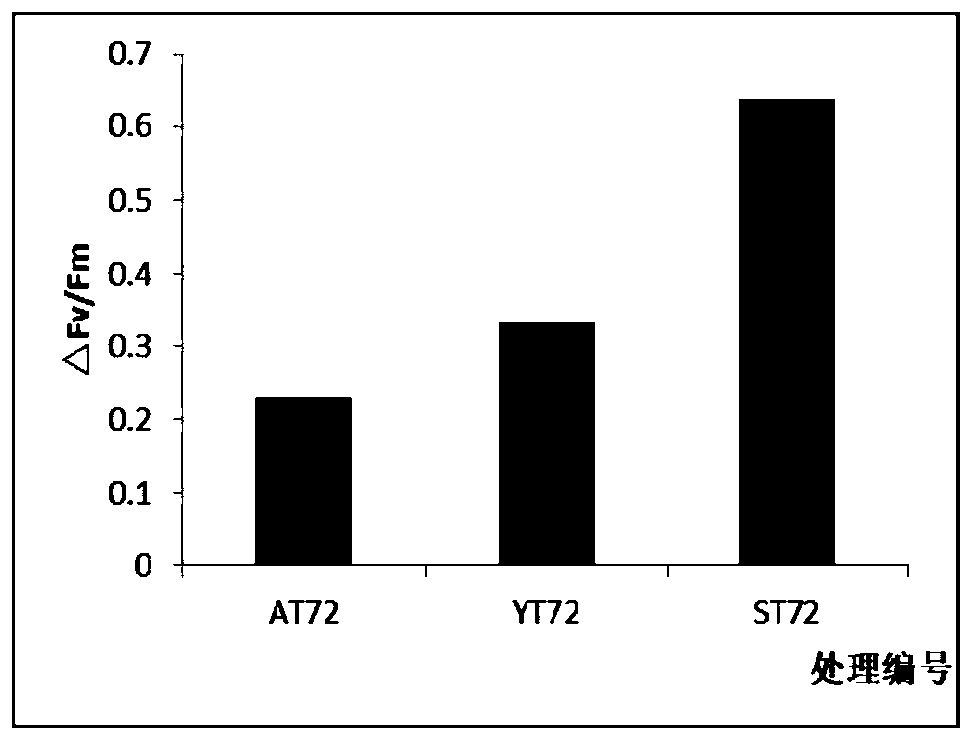
[0040] The △Fv / Fm of bamboo and willow blank control group and salt treatment group after treatment for 72 hours can be seen figure 2 .
[0041] Table 1
[0042]
[0043] The results of observing and measuring the phenotype after different treatments of 0, 24, 48 and 72 of the test materials and the changes of the light energy conversion efficiency Fv / Fm of the total chlorophyll content and the maximum PSII show that: 1) different tree species, different provenances of the same tree species The symptoms caused by the response to salt treatment were different: after 24 hours of salt treatment, a small amount of leaves on the branches of bamboo ...
Embodiment 3
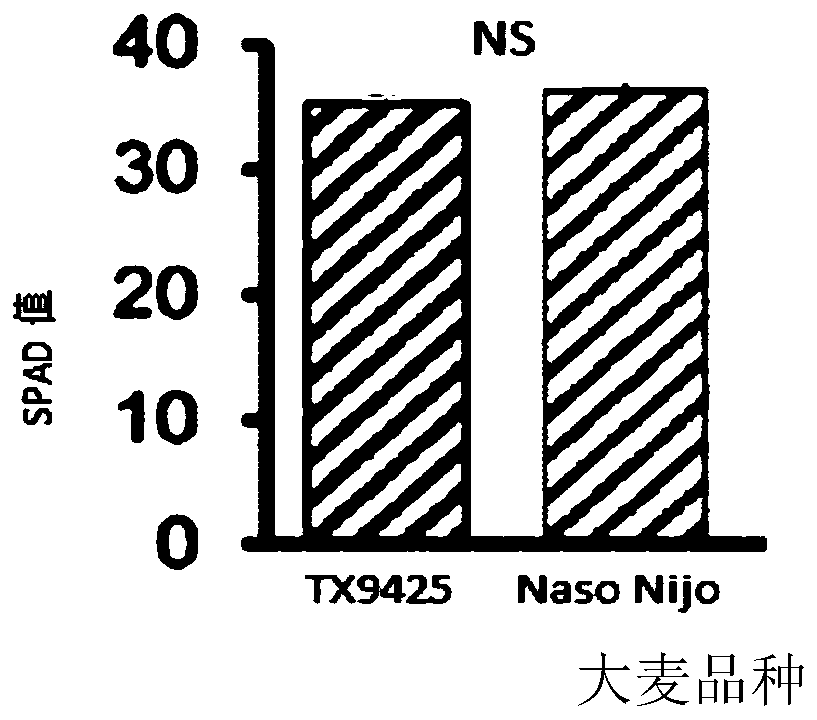
[0045] The detached leaves of the salt-sensitive Naso Nijo barley variety and the salt-tolerant barley variety TX 9425 were treated with water (blank control) and 50mmol / L NaCl solution, respectively, and the two barley varieties were measured after 0h and 48h of the blank control and salt treatment. Varieties of chlorophyll SPAD (relative chlorophyll content) value and CCI (chlorophyll content index) value. The chlorophyll SPAD value is measured by a chlorophyll measuring instrument SPAD-502, calculated according to the amount of transmitted light from the leaves, and the SPAD value is proportional to the absolute content of chlorophyll in the leaves. The chlorophyll content index CCI is measured by a CCM-200 portable chlorophyll meter and calculated by measuring the different absorbance of leaves at two wavelengths of red light and near-infrared light.
[0046] See image 3 ; The results of the chlorophyll SPAD value in the barley leaves in vitro after the blank control (cl...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com