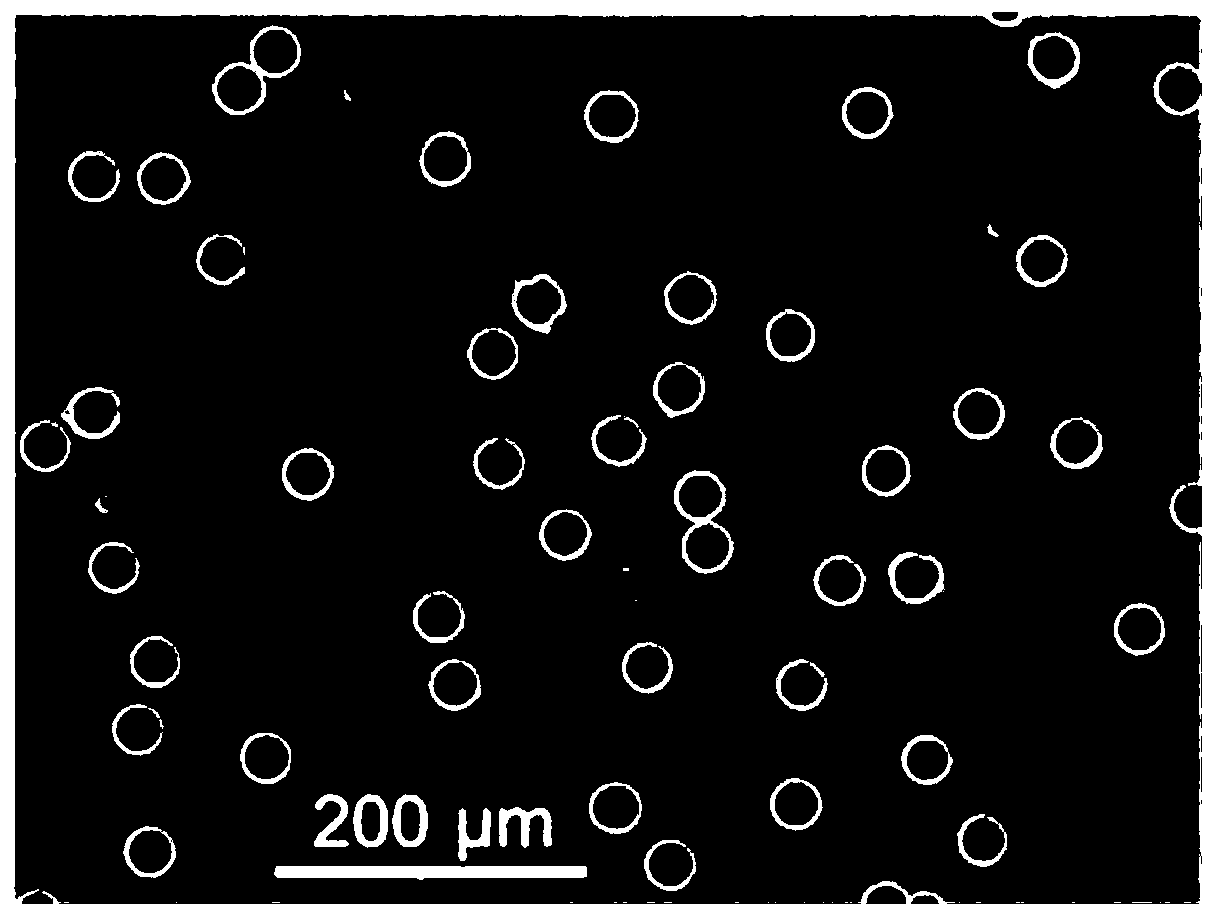
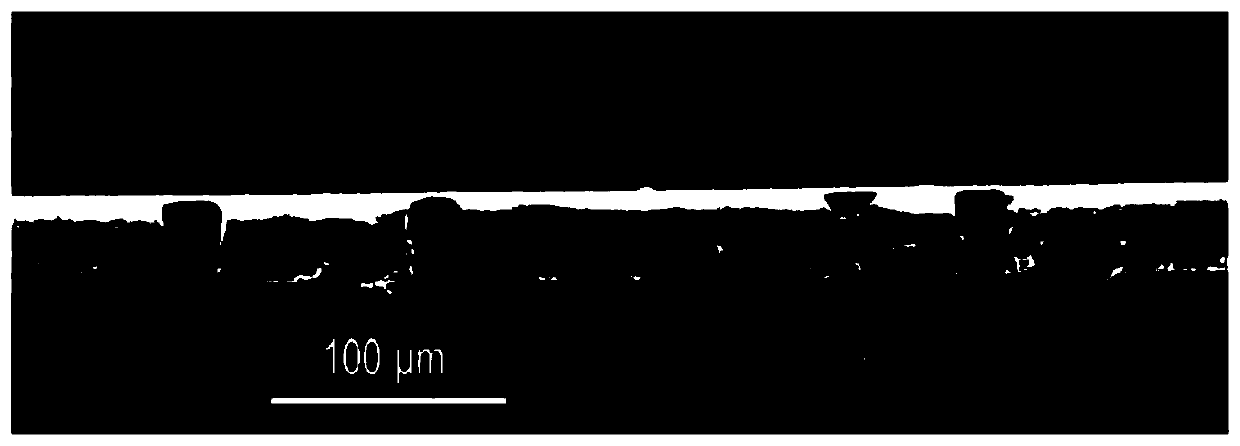
Digital loop-mediated isothermal amplification (LAMP) method based on track etching film and application
A loop-mediated isothermal and isothermal amplification technology, applied in the field of molecular biology, can solve the problems of limited scalability, difficult one-time use, high price, etc., and achieve the effect of flexible and concise operation, simple experimental steps, and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039]Example 1: Analysis of Escherichia coli (ATCC 10798) by a digital loop-mediated isothermal amplification detection method based on a track-etched membrane with a pore size of 25 μm
[0040] The primer sequences used are as follows:
[0041] F3: 5'-GCCATCTCCTGATGACGC-3'
[0042] B3: 5'-ATTTACCGCAGCCAGACG-3'
[0043] LF: 5'-CTTTGTAACAACCTGTCATCGACA-3'
[0044] LB: 5'-ATCAATCTCGATATCCATGAAGGTG-3'
[0045] FIP: 5'-CATTTTGCAGCTGTACGCTCGCAGCCCATCATGAATGTTGCT-3'
[0046] BIP: 5'-CTGGGGCGAGGTCGTGGTATTCCGACAAACACCACGAATT-3'
[0047] The actual original DNA concentrations of Escherichia coli in standard samples a to e are:
[0048] Standard sample a: The original concentration of Escherichia coli DNA is 11copy / μL;
[0049] Standard sample b: the original concentration of Escherichia coli DNA is 1.1×10 2 copy / μL;
[0050] Standard sample c: the original concentration of Escherichia coli DNA is 1.1×10 3 copy / μL;
[0051] Standard sample d: The original concentration of Esc...
Embodiment 2
[0060] Example 2: Analysis of Escherichia coli (ATCC 10798) based on a digital loop-mediated isothermal amplification detection method with a pore size of 14 μm track-etched membrane
[0061] The primer sequences used are the same as those in Example 1, but the actual original concentrations of Escherichia coli DNA in the standard samples a to d in this example are respectively:
[0062] Standard sample a: The original concentration of Escherichia coli DNA is 1×10 3 copy / μL;
[0063] Standard sample b: the original concentration of Escherichia coli DNA is 1×10 4 copy / μL;
[0064] Standard sample c: the original concentration of Escherichia coli DNA is 1×10 5 copy / μL;
[0065] Standard sample d: The original concentration of Escherichia coli DNA is 1×10 6 copy / μL;
[0066] Add 2.5μL standard samples a to e respectively with 1×LAMP buffer, 6mM MgSO 4 , 1.4mM dNTP, 640U / mL Bst DNA polymerase, 1.6μM FIP and BIP, 0.2μM F3 and B3, 0.8μM LF and LB, 1mg / mL BSA, 50μM calcein, 1m...
Embodiment 3
[0073] Example 3 Analysis of Salmonella (CVD 909) by a digital loop-mediated isothermal amplification detection method based on a track-etched membrane with a pore size of 25 μm
[0074] The primer sequences used are as follows:
[0075] F3: 5'-GACTTGCCTTTAAAAGATACCA-3'
[0076] B3: 5'-AGAGTGCGTTTGAACACTT-3'
[0077] LF: 5'-TCGGATGGCTTCGTTCCT-3'
[0078] LB: 5'-CAAGGGTTTCAAGACTAAGTGGTTC-3'
[0079] FIP: 5'-AACTTGCTGCTGAAGAGTTGGACCGAATGACTCGACCATC-3' BIP: 5'-CCTGGGGCCAAATGGCATTATGCACTAAGTAAGGCTGG-3'
[0080] The actual original DNA concentrations of Salmonella in standard samples a to e are:
[0081] Standard sample a: the original concentration of Salmonella DNA is 11copy / μL;
[0082] Standard sample b: the original concentration of Salmonella DNA is 1.1×10 2 copy / μL;
[0083] Standard sample c: the original concentration of Salmonella DNA is 1.1×10 3 copy / μL;
[0084] Standard sample d: the original concentration of Salmonella DNA is 1.1×10 4 copy / μL;
[0085] Stan...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
| pore size | aaaaa | aaaaa |
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com