A model construction method and application for identifying tumor purity samples
A construction method and tumor technology, which is applied in the field of model construction for identifying tumor purity samples, can solve problems such as difficulty in estimating tumor purity, fewer mutation sites in chip captured data, etc., and achieve high reliability results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 Construction of the identification model
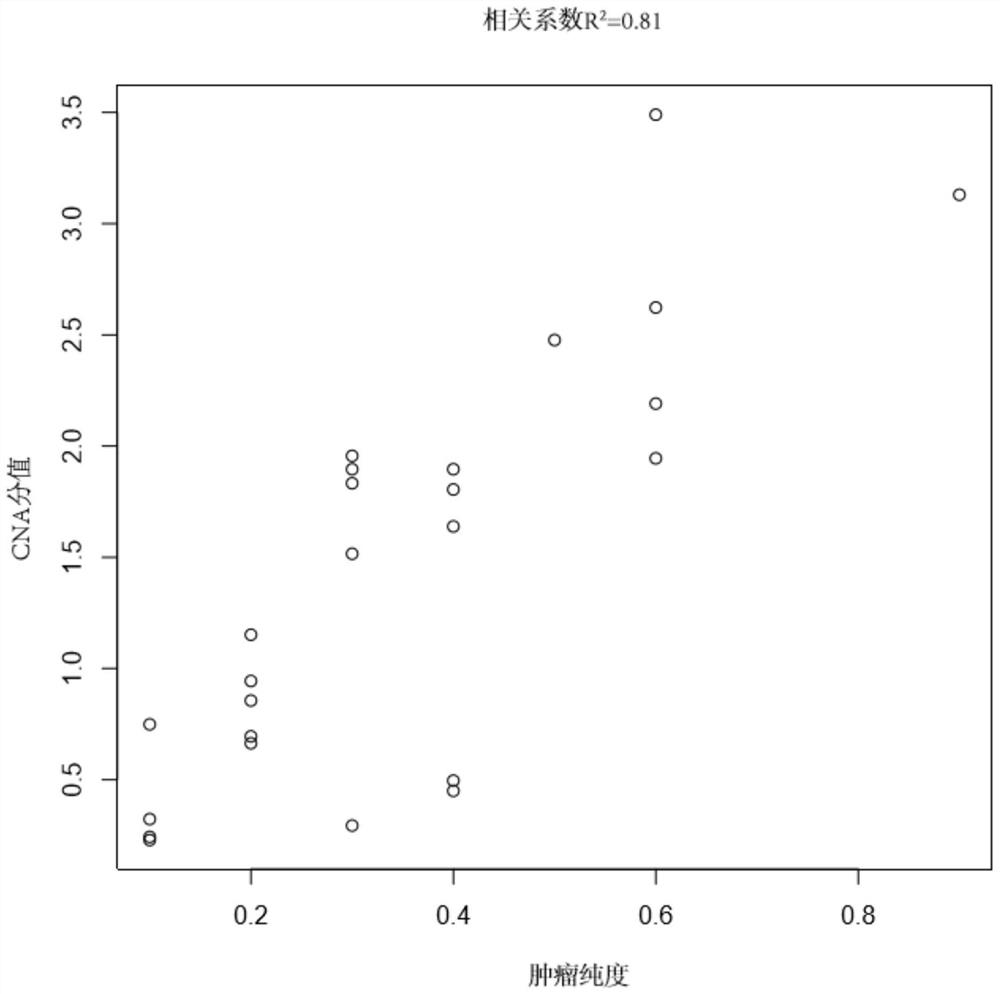
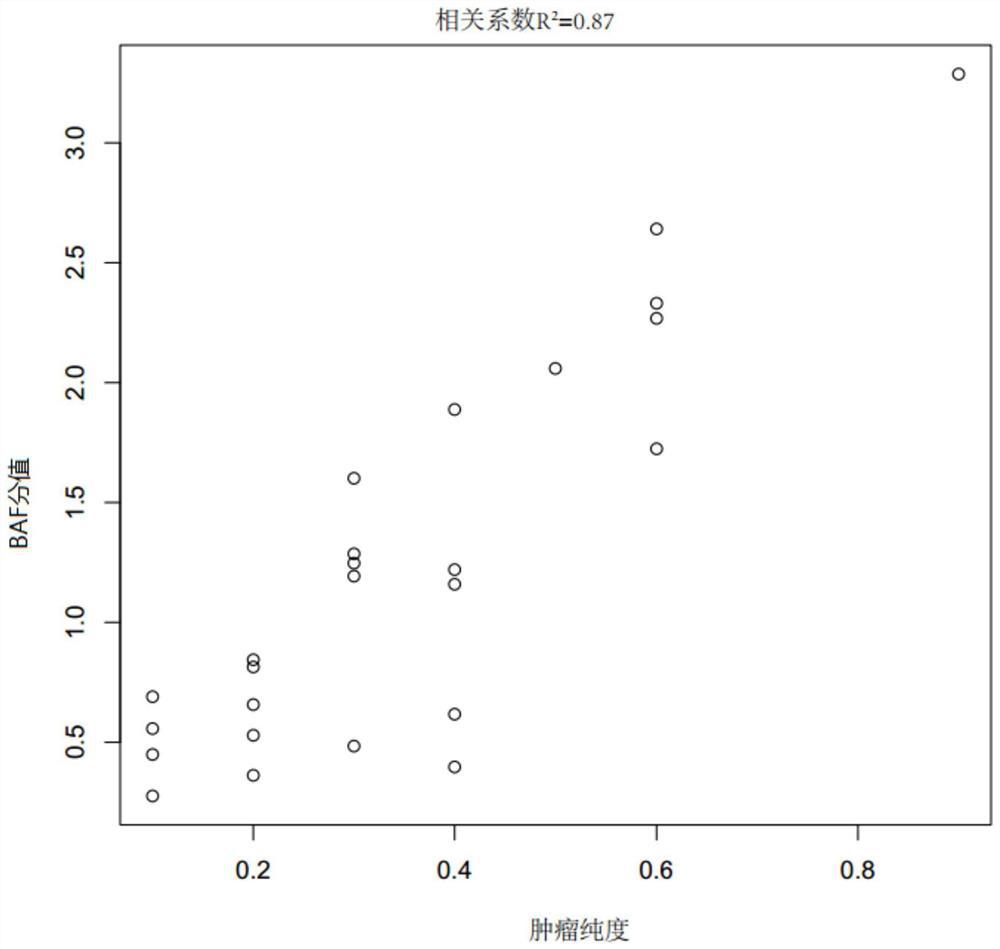
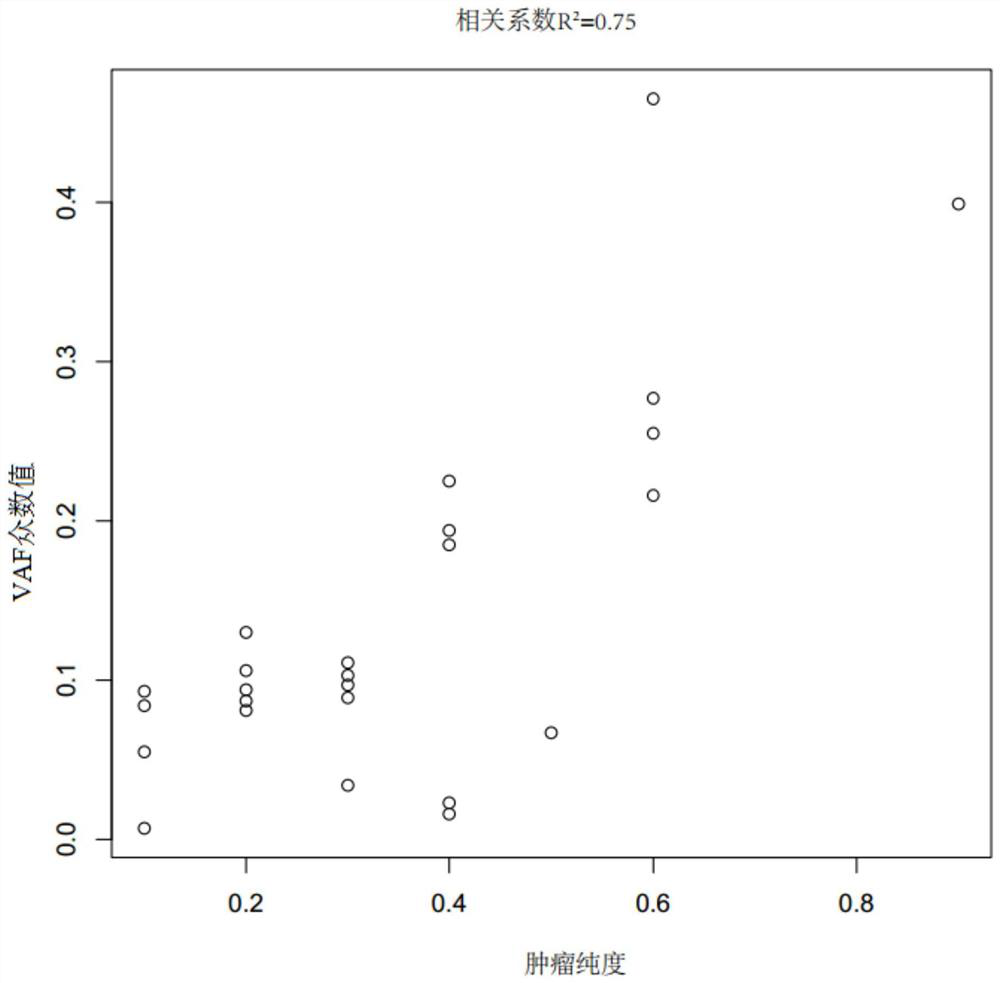
[0044] (1) Take 40 microscopic examination results (estimated tumor purity by image analysis of hematoxylin and eosin stained sections) as tumor samples of known tumor purity and their paired paracancerous tissues (paired samples) ), see Table 1-2 for details. The target probes (Beijing Gene Plus Technology Co., Ltd. OncoD-C1021 tumor drug gene detection product, OncoD-P1021 tumor drug gene detection product and OncoMRD tumor postoperative monitoring product) were captured and sequenced respectively, and the sequencing platform was Illumina NovaSeq sequencing. The data includes the sequencing data of the target capture region (on target) and the non-target capture region (off target). The above sequencing data is GC-corrected using the LOESS algorithm to the target capture sequencing data, and then compared with the reference genome ( Human reference genome, USCS hg19) is compared to obtain the comparison result file...
Embodiment 2
[0052] Example 2 Identification of tumor purity
[0053] Select tumor tissue samples A (No.: 180028560F) and B (No. 180027591F) that have undergone microscopy and the results of microscopy (estimated tumor purity by image analysis of hematoxylin and eosin-stained sections) as known purity. And its paired adjacent tissue (numbering 190004260 BCD and 180027591 BCD), according to the step (1)-step (4) in the embodiment 1 implementation, the obtained index data CNA score, BAF score and VAF mode value are shown in the following table 3.
[0054] Table 3 Data of each indicator
[0055]
[0056] In this example, it is assumed that the purity is < 0.4 (40%) as low tumor purity, and the purity ≥ 0.4 (40%) is considered as high tumor purity.
[0057] When single-index identification is adopted, any one of the index data CNA score, BAF score and VAF mode value in Table 3 is substituted into the corresponding linear model in step (5) in Example 1, and the tumor purity can be obtained...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com