Application of a kind of complement serine protease
A serine protease and complement technology, applied in the field of molecular biology, can solve the problems of unreported functions, no enlightenment, unresearched functions and applicability of complement I factor or serine protease, etc., and achieves the effects of no potential safety hazards and simple preparation process.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Preparation of Serine Protease Recombinant Protein of Complement Regulating Protein I Factor (1) Construction of Serine Protease Expression Plasmid pEtPoCFI-Tryp The flounder complement serine protease of the present invention is obtained by prokaryotic protein expression system, and its sequence is in SEQ ID No. 1 of the sequence table amino acid sequence.
[0027] Sequence Listing SEQ ID No.1 is:
[0028] RVVGGVPAKPTQIQWQIALEENKKIDCGGAYIGGCWVLTAAHCVRPNPVPFKVKFSLWRKWSAQDTTDIVPVEDIRIHPKYNAATYENDIALVKLEKLPFKDKCFEDNPAISAVCVPWSTQLFQANHTCSISGWGRTIDGRAAQVLLWANVSLIDNCQRFYKDRFRPGMMCAG
[0029] (a) Sequence features:
[0030] ●Length: 173
[0031] ●Type: amino acid sequence
[0032] ●Chain type: single chain
[0033] ●Topology: Linear
[0034] (b) Molecular type: protein
[0035] (c) Assumption: No
[0036] (d) Antonym: No
[0037] (e) The original source: the flounder cDNA was used as the template, and the primers F1 and R1 were used for PCR amplification. The PCR prod...
Embodiment 2
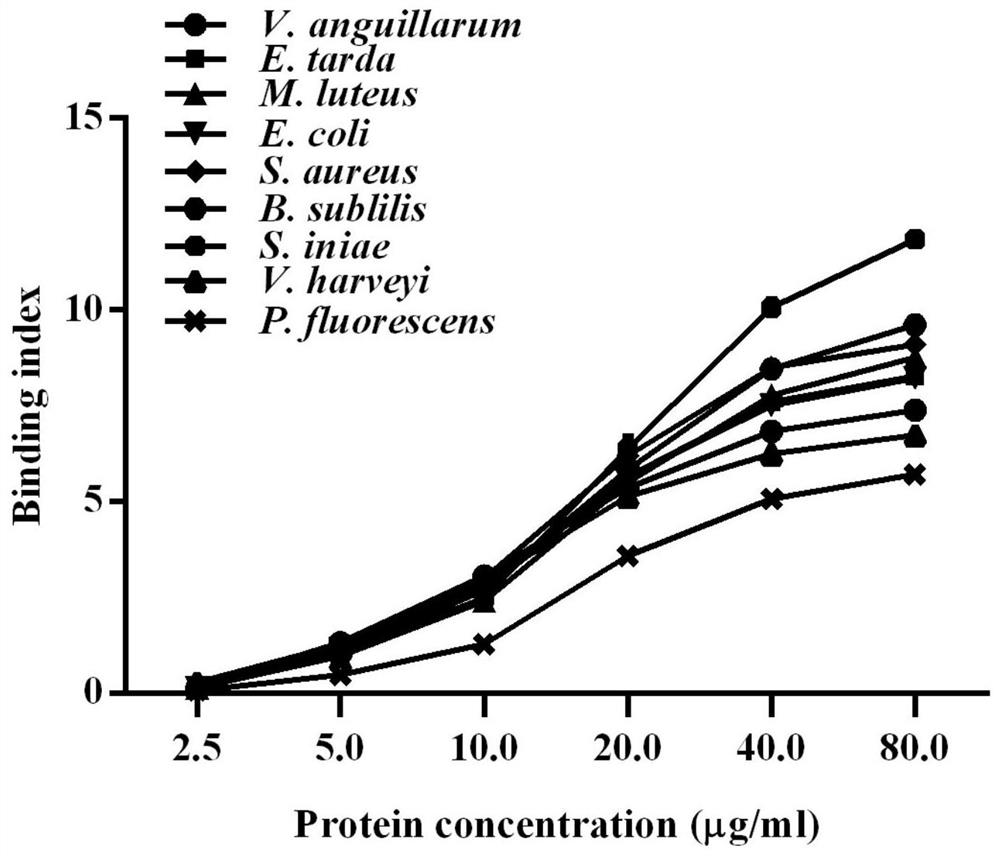
[0044] Detection of the ability of recombinant protein containing serine protease (rPoCFI-Tryp) to bind to various bacteria: different bacteria were inoculated in 5ml liquid LB medium and cultured to OD 600About 0.8, among them, Edwardella lentus, Vibrio harveii, Vibrio eel, and Pseudomonas fluorescens were cultured at 28°C; Escherichia coli, Micrococcus luteus, Staphylococcus aureus, and Bacillus subtilis were cultured at 37°C nourish;
[0045] Inoculate Streptococcus iniae in TSB medium at 28°C to OD 600 about 0.8.
[0046] Dilute each of the above bacteria with coating solution to 10 8 CFU / ml, as bacterial dilution, the purified protein in Example 1 above was diluted in PBS to 2.5 μg / ml, 5 μg / ml, 10 μg / ml, 20 μg / ml, 40 μg / ml, 80 μg / ml, namely Diluent of rPoCFI-Tryp.
[0047] The above bacterial dilutions were mixed with various concentrations of dilutions of the serine protease rPoCFI-Tryp or the tagged protein rTrx (control) and added with CaCl at a final concentration...
Embodiment 3
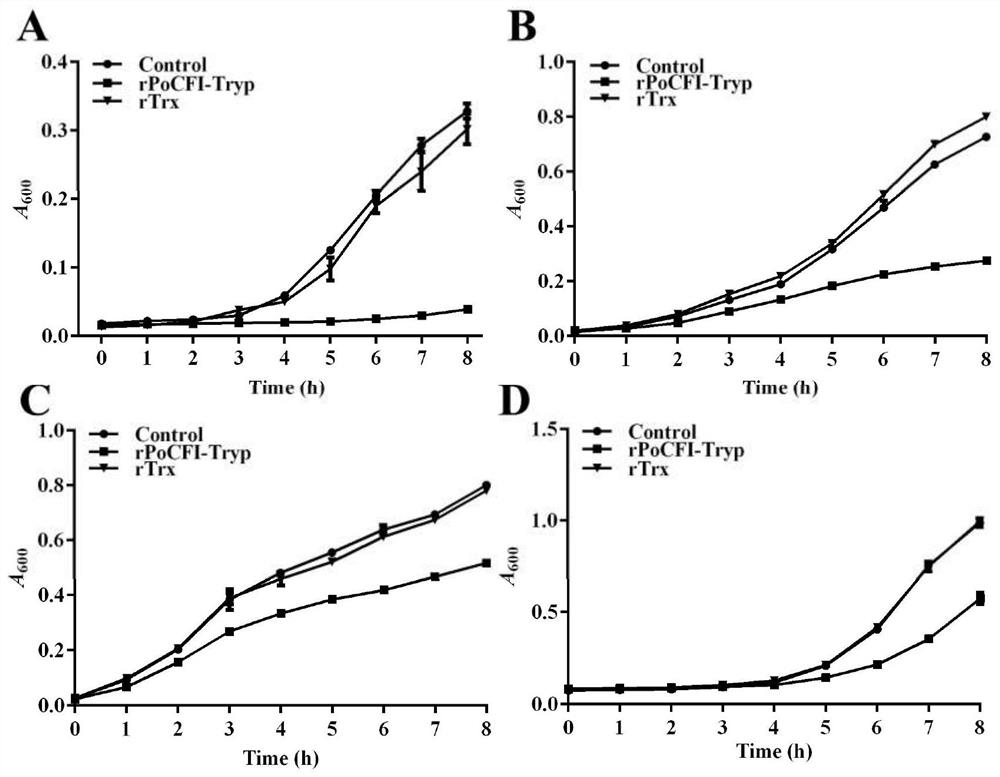
[0051] The ability of serine protease rPoCFI-Tryp to inhibit the growth of various bacteria Step 1) Culture of bacteria
[0052] Edwardsiella lentus, Vibrio harveii, and Vibrio eel were inoculated in 5ml of liquid LB; Streptococcus iniae was inoculated in TSB medium, and each strain was cultured to OD in their respective medium in a conventional manner 600 about 0.8, and then the culture medium was diluted to 10 with each new medium. 6 CFU / ml. Step 2) Determine the growth curve of different bacteria
[0053] The purified protein in the above Example 1 was diluted to 70 μg / ml in PBS, which was the rPoCFI-Tryp dilution solution. The above bacterial dilution solutions were mixed with the serine protease rPoCFI-Tryp dilution solution or the tagged protein rTrx (control group), and added. CaCl at a final concentration of 50 μM 2 , as for culturing in a 28°C incubator, measure the absorbance at 600 nm every hour, and draw the growth curves of different bacteria (see figure 2 )....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com