Method for cultivating soft rice by using transcriptional activation factor sample effector nuclease technology
A transcription activator and effector technology, applied in the field of soft rice with low amylose content, can solve the problems of large randomness, long time, non-specific mutations, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Example 1: Identify rice OsGBSSI Synthesis of the functional module TN12a at the left target site of the gene
[0018] The TN12a module was synthesized by gene synthesis method (Nucleic Acids Research, 2004, 32, e98). The designed primers are:
[0019] TN12a -1 GGATCCCTCACTCCAGCACAGGTGGTTGCGATCGCA TCCA ATATCGGAGG TAAGCAGGCG (shown in SEQ ID NO. 3)
[0020] TN12a -2 CCTGACAAAGGACCGGCAACAGACGCTGCACAGTC T CCAA CGCCTGCTTA CCTCCGATAT (shown in SEQ ID NO.4)
[0021] TN12a -3 GTTGCCGGTCCTTTGTCAGGATCATGGCCTGACGC CAGAT CAGGTAGTCG CAATCGCAAG (shown in SEQ ID NO.5)
[0022] TN12a -4 GCGCTGGACTGTCTCAAGGGCCTGCTTGCCTCCAT TGTTG CTTGCGATTG CGACTACCTG (shown in SEQ ID NO.6)
[0023] TN12a -5 CCCTTGAGACAGTCCAGCGCTTGTTGCCTGTTCTAT GCCA GGATCACGGC CTGACTCCTG (shown in SEQ ID NO. 7)
[0024] TN12a -6 TGTTTACCACCATCGTGAGACGCAATGGCGACCACT TGGTCAGGAGTCAG GCCGTGATCC (shown in SEQ ID NO. 8)
[0025] TN12a -7 TCTCACGATGGTGGTAAACAAGCACTGGAAACGGT TCAAA GACTCCTCCC AGTGCTGTGT (shown in SEQ ID NO.9)
[0026] TN12...
Embodiment 2
[0058] Example 2: Identify rice OsGBSSI Synthesis of the functional module TN12b at the right target site of the gene
[0059] Using gene synthesis method (Nucleic Acids Research, 2004, 32, e98) TN12b. The designed primers are:
[0060] TN12b-1 GGATCCCTCACTCCAGCACAGGTGGTTGCGATCGCA TC CAATATCGGAGGTAAGCAGGCG (shown in SEQ ID NO.41)
[0061] TN12b -2CCTGACAAAGGACCGGCAACAGACGCTGCACAGTCTC CAACGCCTGCTTACCTCCGATAT (shown in SEQID NO.42)
[0062] TN12b-3 GTTGCCGGTCCTTTGTCAGGATCATGGCCTGACGCCA G ATCAGGTAGTCGCAATCGCAAG (shown in SEQ ID NO. 43)
[0063] TN12b-4 GCGCTGGACTGTCTCAAGGGCCTGCTTGCCTCCATTA TTGCTTGCGATTGCGACTACCTG (shown in SEQ ID NO.44)
[0064] TN12b-5 CCCTTGAGACAGTCCAGCGCTTGTTGCCTGTTCTATGC CAGGATCACGGCCTGACTCCTG (shown in SEQ ID NO.45)
[0065] TN12b-6 TGTTTACCACCGATGTTAGACGCAATGGCGACCACTT GGTCAGGAGTCAGGCCGTGATCC (SEQID NO.46)
[0066] TN12b-7TCTAACATCGGTGGTAAACAAGCACTGGAAACGGTTCAAAGACTCCTCCCAGTGCTGTGT (shown in SEQID NO.47)
[0067] TN12b-8 CTATAGCTACGACTTGTTCAGGTGTCAAACCATGATC TTGACACAG...
Embodiment 3
[0099] Example 3: Targeting GBSSI Construction of TALEN Plant Expression Vector of Gene
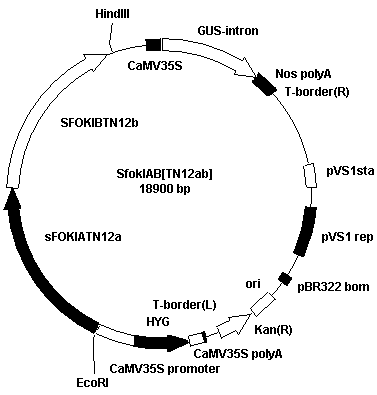
[0100] The left target site recognition module pUC19 (TN12a) and the right target site recognition module pUC19 (TN12b) of GBSSI gene were double digested with BamHI and SacI. The digested products were separated by Agarose electrophoresis and gel tapping to recover 1500bp fragments. The SFokIA (pUC19) vector and TN12a fragment were ligated with T4 DNA ligase; the SFokIB (pUC19) vector and TN12b fragment were ligated with T4 DNA ligase. The above linkers were transformed into Escherichia coli DH5α, single colonies of Escherichia coli transformants were picked for liquid culture, the plasmids were extracted and the enzyme digestion was carried out, and finally the full sequence analysis and determination of the inserts in the positive plasmids were performed to obtain the targeted GBSSI The two TALEN plasmids of the gene are named SFokIA【TN12a】pUC19 and SFokIB【TN12b】pUC19.
[0101] The SFokIA...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com