Method and system for finding molecular marker based on transcriptome time sequence dynamic change and gene function association
A molecular marker and gene function technology, applied in the field of finding biomarkers, can solve problems such as difficulty in obtaining project approval or investment, high verification risk, and large experimental volume, achieving low requirements for parameter optimization of measurement accuracy and methods, and successful verification. High rate and robust effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
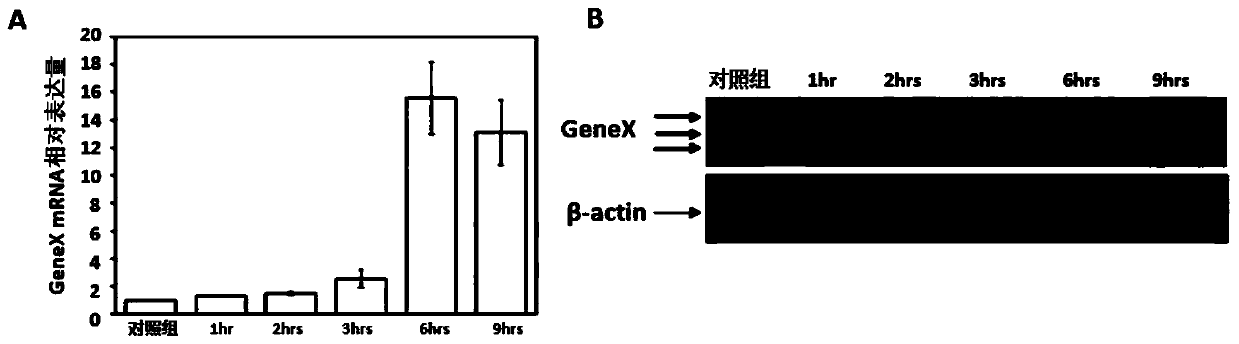
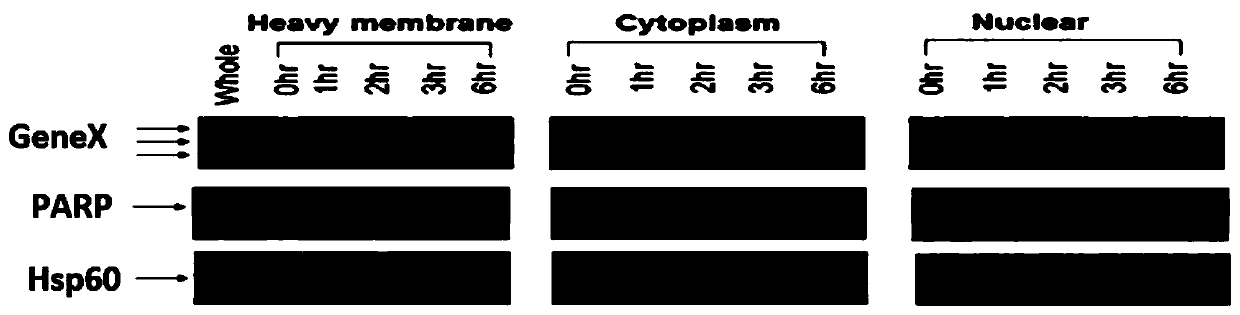
Examples
Embodiment Construction
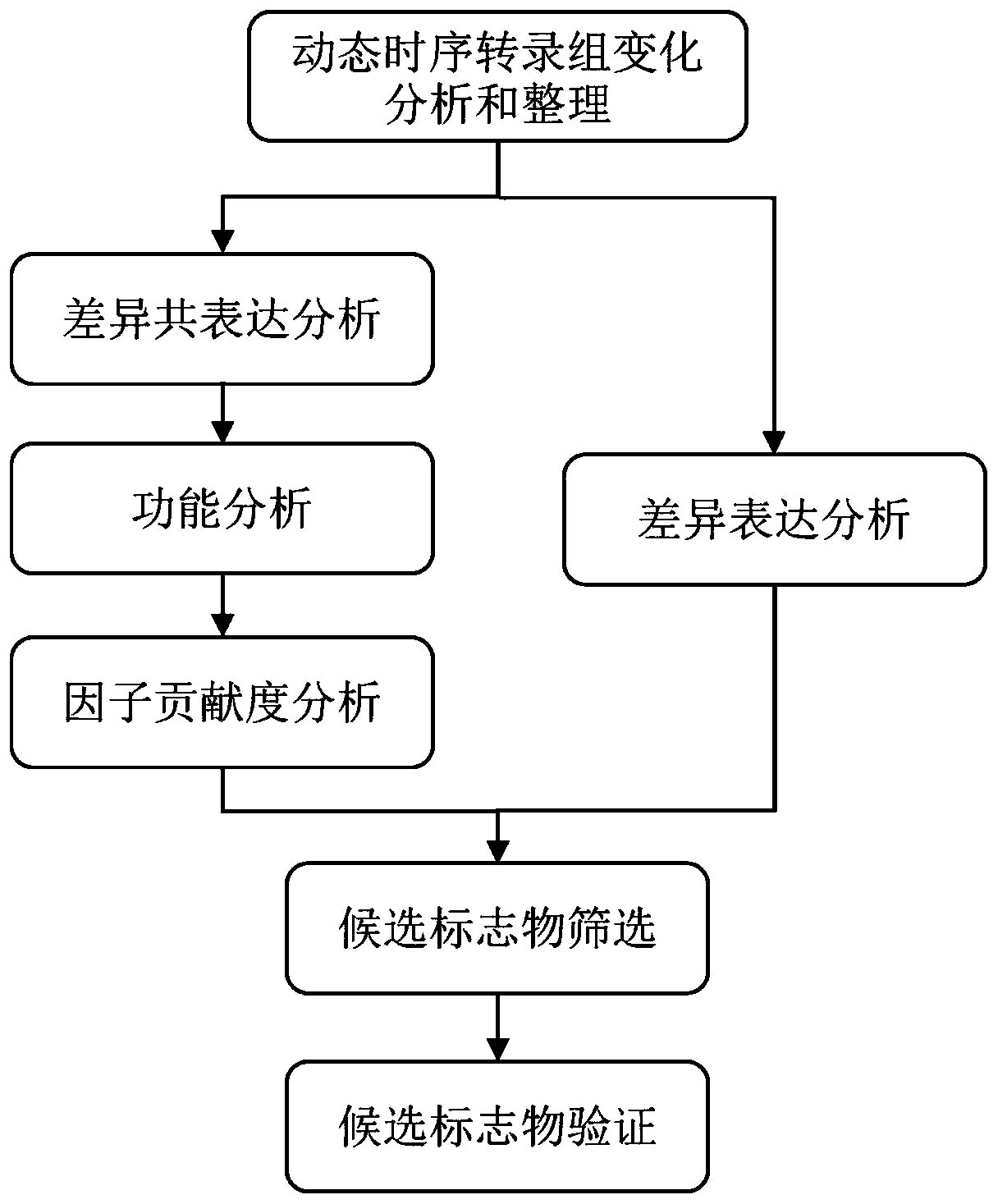
[0058] The invention discloses a system, method and application for discovering molecular markers based on the dynamic changes of transcriptome time series and gene function correlation. The method provided by the invention can be realized by the following systems:
[0059] Dynamic temporal transcriptome change analysis and sorting module to obtain mRNA expression data, including target pathophysiological processes and control pathophysiological processes;
[0060] The differential co-expression analysis module is used to perform differential co-expression analysis on mRNA data to find gene sets that are co-expressed in the target pathophysiological process and have no expression correlation in the control pathophysiological process;
[0061] A functional analysis module for finding gene sets functionally related to the target pathophysiological process on the gene function association network;
[0062] The factor contribution analysis module is used to evaluate the associatio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com