Method for breeding high-yield shiny-leaved yellowhorn strain
A high-yield and high-yield technology for Fructus Fructus, applied in the fields of botanical equipment and methods, plant genetic improvement, cultivation, etc., can solve problems such as the failure to identify the method for self-crossing, fertile, high-yield and excellent plants of Fructus Fructus and the cultivation technology of new varieties.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Embodiment 1, the acquisition of X. sorbifolium genomic DNA
[0064] The genomic DNA of the sample was extracted by the CTAB method, and the specific operation was as follows:
[0065] 1) Preheat the CTAB extract in a 65°C water bath;
[0066] 2) Grind the sample quickly in liquid nitrogen, transfer the powdered material into a 2mL centrifuge tube, add preheated CTAB extract (3-5ml of extract per gram of sample), and keep warm at 65°C for 30-60min, every Gently invert and mix for 10 minutes;
[0067] 3) Centrifuge at 11000rpm for 5min, take the supernatant and transfer it to a new centrifuge tube;
[0068] 4) Add an equal volume of phenol / chloroform (1:1, volume ratio), mix thoroughly, centrifuge at 11,000 rpm for 10 min, and transfer the supernatant to a new centrifuge tube;
[0069] 5) Add an equal volume of chloroform, mix thoroughly, centrifuge at 11000rpm for 10min, and transfer the supernatant to a new centrifuge tube;
[0070] 6) Repeat steps 4) and 5);
[0...
Embodiment 2
[0076] Embodiment 2, SSR primer screening
[0077] (1) PCR reaction system:
[0078] SSR (total 20 μL): ddH 2 7.2 μL of O, 10 μL of MIX, 0.3 μL of forward primer F, 0.3 μL of reaction primer R, 2 μL of DNA template (genomic DNA of X. sorbifolium obtained in Example 1), and 0.2 μL of Taq.
[0079] (2) The PCR reaction adopts the following cycle parameters:
[0080] SSR PCR amplification program: pre-denaturation at 94°C for 5 minutes; denaturation at 94°C for 30 s, annealing at 54°C for 35 s, extension at 72°C for 40 s, a total of 35 cycles; final extension at 72°C for 3 min.
[0081] (3) Primer information:
[0082] Table 1 Screening primer information
[0083]
[0084]
[0085] (4) The PCR product was added with loading buffer, denatured at 94° C. for 10 min, then analyzed by vertical electrophoresis on a 6% denaturing polyacrylamide gel, and observed after silver staining. Partial picture of primer screening in denaturing polyacrylamide gel electrophoresis (PAGE) ...
Embodiment 3
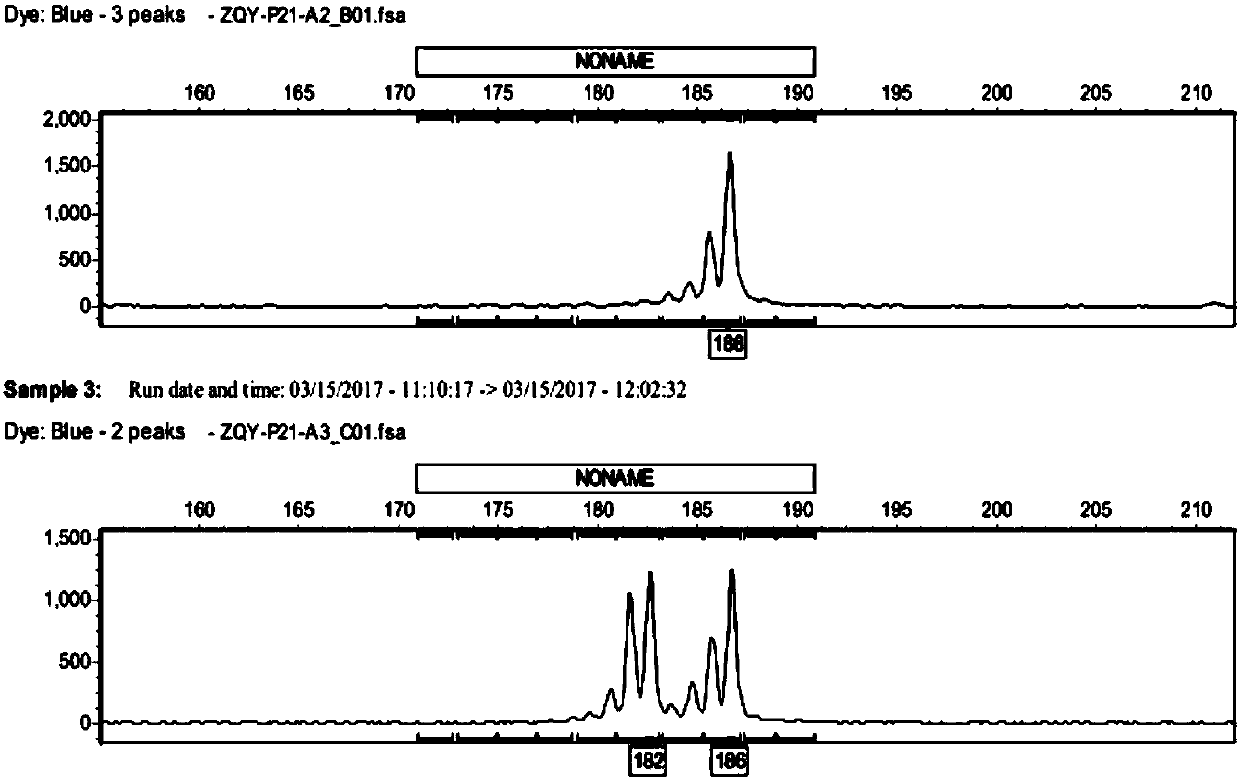
[0086] Embodiment 3, detection by capillary electrophoresis
[0087] Using the 19 pairs of SSR primers (Table 1) with clear bands, high polymorphism and good repeatability screened in Example 2 above, fluorescent primers were synthesized, and the fragment size was detected by capillary electrophoresis analysis.
[0088] 1. PCR amplification
[0089] (1) PCR reaction system:
[0090] SSR fluorescent primer system (total 20 μL): ddH 2 O 14.8 μL, dNTP 0.4 μL, Buffer 2 μL, forward primer F 0.3 μL (20 μM), reverse primer R 0.3 μL (20 μM), DNA template 2 μL, Taq 0.2 μL.
[0091] (2) The PCR reaction adopts the following cycle parameters:
[0092] SSR PCR amplification program: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 s, annealing at 54°C (annealing temperature fluctuates around 54°C) for 35 s, extension at 72°C for 40 s, a total of 35 cycles; final extension at 72°C for 3 min.
[0093] 2. Capillary electrophoresis analysis
[0094] After mixing the formam...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com