Method and special culture medium for SILAC labelling of mycobacterium smegmatis protein
A technology of Mycobacterium smegmatis and culture medium, applied in the directions of microorganism-based methods, biochemical equipment and methods, bacteria, etc., can solve the problem of unknown effect, unsatisfactory labeling effect of Mycobacterium smegmatis, negative effects of labeling effect, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
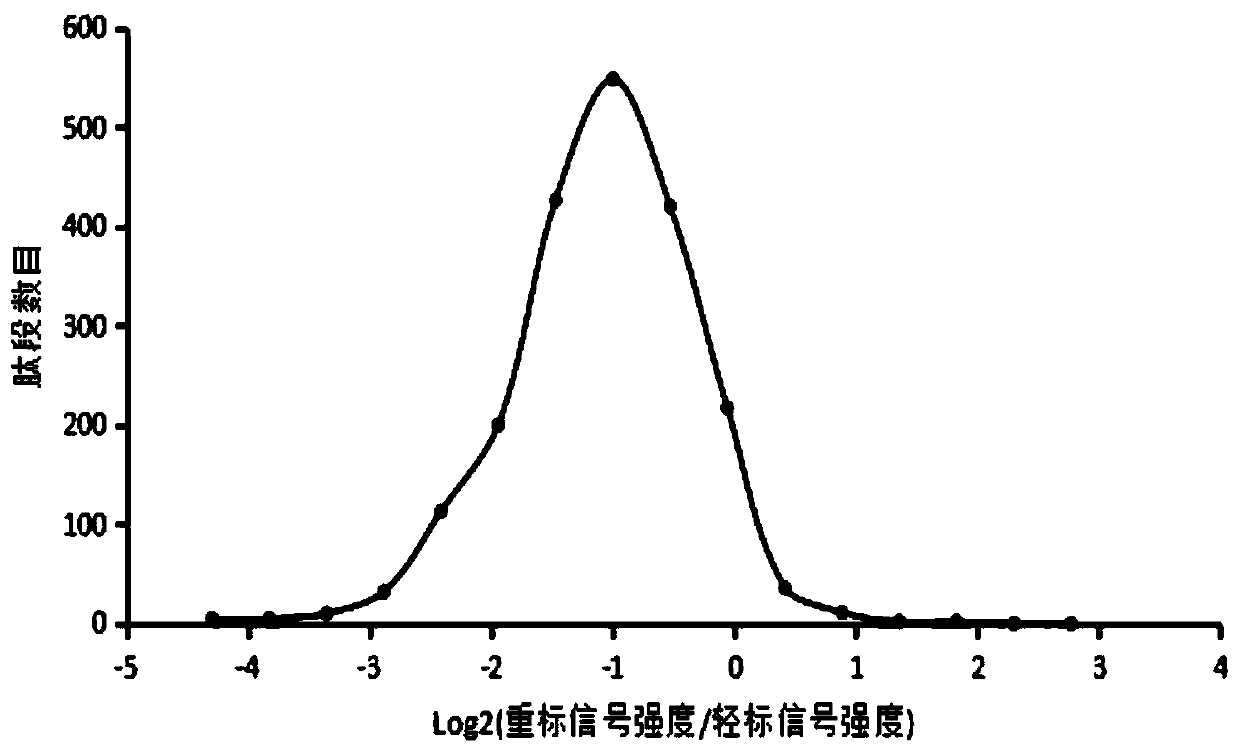
[0058] Example 1, Mycobacterium smegmatis proteome marker efficiency test in efficient prokaryote SILAC marker medium
[0059] The inventors firstly used the full marker medium (Ping L, Zhang H, Zhai L, Dammer EB., Duong DM., Li N, Yan Z, Wu J, XuP. Quantitative proteomics reveals significant changes in cell shape and anenergy shift after IPTG induction via an optimized SILAC approach for Escherichia coli.J Proteome Res.2013,12:5978-5988.) on Mycobacterium smegmatis M.smegmatisMC 2 155 conducted SILAC labeling studies. The Escherichia coli SILAC marker medium SILACE used by the inventors is shown in Table 1.1.
[0060] Table 1.1 Components of SILACE special medium for SILAC-labeled Escherichia coli
[0061]
[0062]
[0063] 1.1. Preparation of SILAC-labeled E. coli medium SILACE
[0064] According to the above-mentioned components and their final concentrations, configure with ultrapure water to obtain Escherichia coli SILAC labeling medium SILACE. Inorganic salts a...
Embodiment 2
[0075] Embodiment 2, the design and labeling efficiency test of Mycobacterium smegmatis SILAC labeling medium ZJL-x
[0076] Since Mycobacterium smegmatis did not achieve high labeling efficiency in the medium SILACE, we chose Middlebrook 7H9, a commonly used medium for mycobacteria [Atlas, Ronald M.; James W. Snyder (2006). Handbook of media for clinical microbiology.CRC Press.ISBN 978-0-8493-3795-6.] as the basal medium (the composition of the basal medium is shown in Table 2.0), and the gluten that affects the biosynthesis of lysine and arginine Amino acid and aspartic acid were removed, and Mycobacterium smegmatis-specific SILAC marker medium ZJL-x was designed, the components of which are shown in Table 2.1.
[0077] Table 2.0 Composition of Mycobacterium smegmatis basal medium Middlebrook 7H9
[0078] components Final concentration (mg / L) ammonium sulfate 500 L-Glutamic Acid (L-Glutamic Acid) 500 Pyridoxine 1 biotin 0.5 Sodium ci...
Embodiment 3
[0097] Embodiment 3, the design and labeling efficiency test of Mycobacterium smegmatis SILAC special marker medium ZJL-y
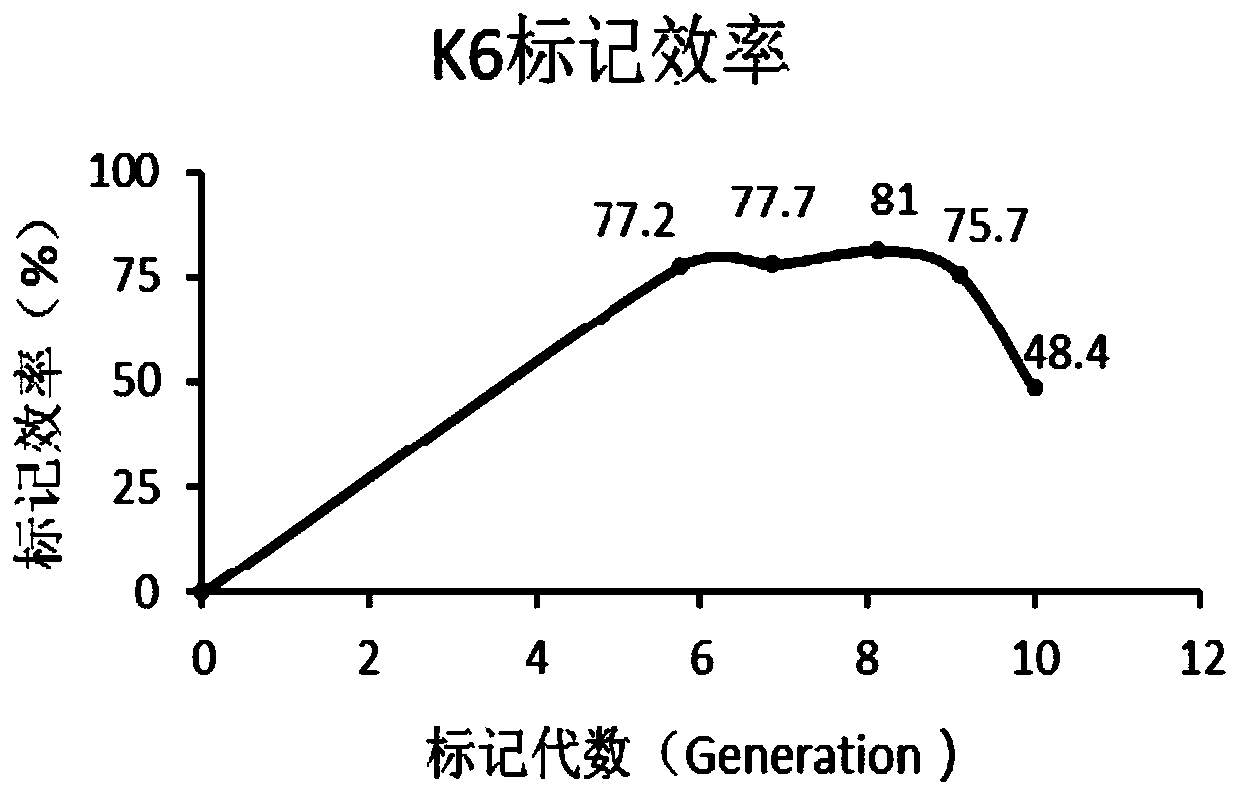
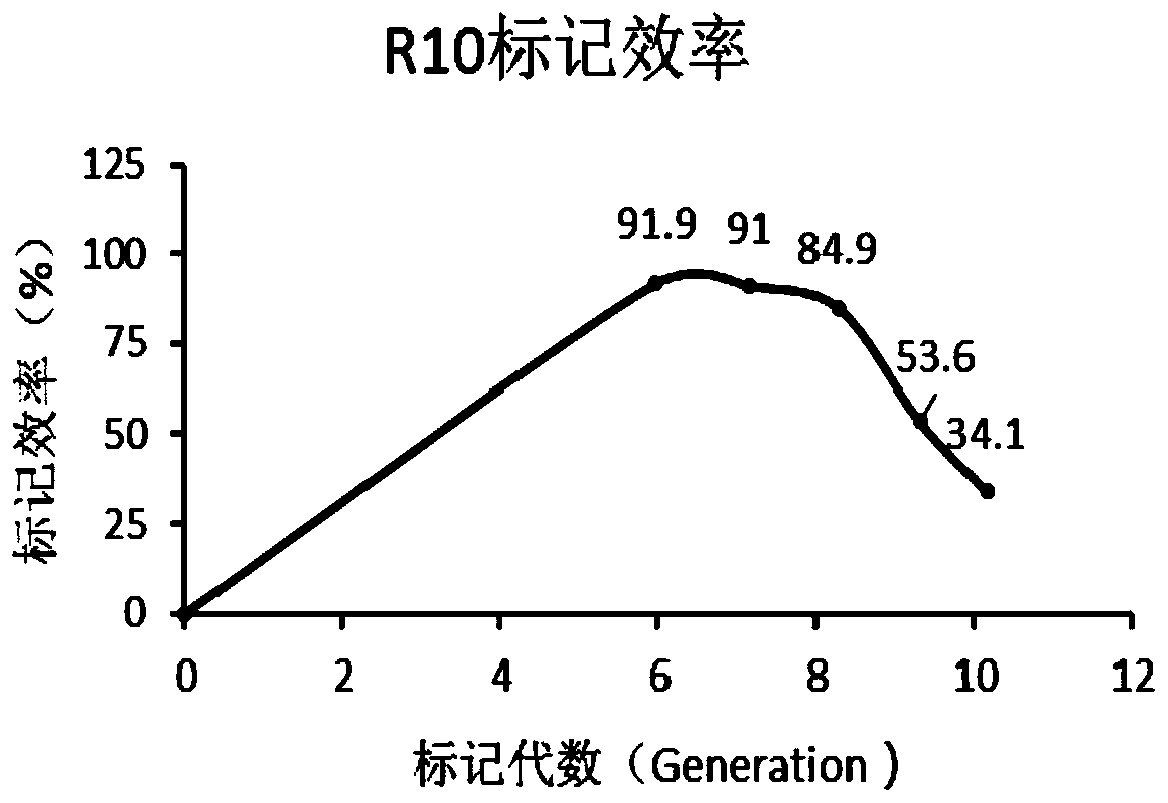
[0098] Based on the results of Example 2, we doubled the content of heavy-labeled amino acids K6 and R10 on the basis of the medium ZJL-x, and designed the Mycobacterium smegmatis SILAC labeling medium ZJL-y as shown in Table 3.1.
[0099] Table 3.1 Components of ZJL-y special medium for SILAC-labeled Mycobacterium smegmatis
[0100]
[0101]
[0102] 3.1. Configuration of Mycobacterium smegmatis SILAC marker medium ZJL-y
[0103] According to the above-mentioned components of ZJL-y and its final concentration, it was configured with ultrapure water to obtain a culture medium for the SILAC labeling experiment of Mycobacterium smegmatis. Inorganic salts and glucose are sterilized by autoclaving, and amino acids are sterilized by filtration.
[0104] 3.2. Extraction of total protein
[0105] The 4th, 5th, 7th, and 8th generation SILAC-labeled M.sme...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com