Design synthesis method of spider silk protein and spinning
A technology of spidroin and protein, applied in the field of biomedical materials, can solve the problems of fiber inhomogeneity, low solubility of recombinant spidroin, difficulty in spinning, etc., and achieve the effect of uniform thickness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1: Spinning spider silk in a pH 2 coagulation bath
[0062] 1. Construction of recombinant expression vectors and strains
[0063] ①Construction of recombinant expression vector
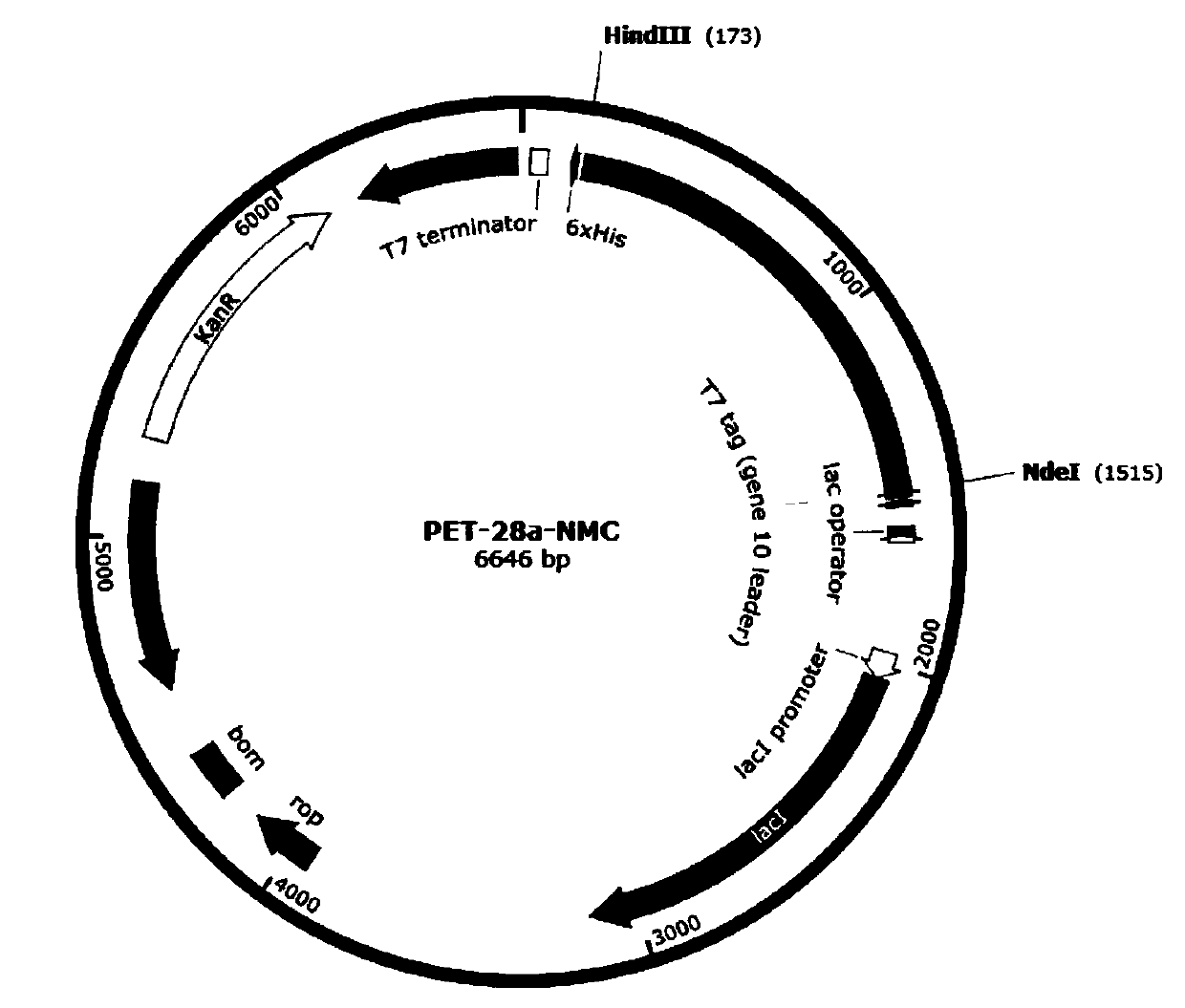
[0064]The N-terminal domain (SEQ ID NO.1) of E.australis spider MaSp1 protein, the C-terminal domain (SEQ ID NO.2) of A.ventricosus spider and the core domain (SEQ ID NO.3) of C.moluccensis origin ) is fused by genetic engineering technology, and codon optimization is performed on the gene according to the expression preference of Escherichia coli without changing its amino acid sequence (SEQ ID NO.4). A NdeI restriction site was added at the 5' end of the gene sequence and a HindIII restriction site was added at the 3' end of the gene. The gene sequence (SEQ ID NO.5) was synthesized and cloned into the Escherichia coli expression vector pET-28a to obtain a recombinant expression vector pET-28a-NMC (SEQ ID NO.6), which contained a strong T7 promoter , lca lactose operon, kanamycin r...
Embodiment 2
[0085] Example 2: Spinning spider silk in pH 3 coagulation bath
[0086] 1. Construction of recombinant expression vectors and strains
[0087] ①Construction of recombinant expression vector
[0088] The N-terminal domain (SEQ ID NO.1) of E.australis spider MaSp1 protein, the C-terminal domain (SEQ ID NO.2) of A.ventricosus spider and the core domain (SEQ ID NO.3) of C.moluccensis origin ) is fused by genetic engineering technology, and codon optimization is performed on the gene according to the expression preference of Escherichia coli without changing its amino acid sequence (SEQ ID NO.4). A NdeI restriction site was added at the 5' end of the gene sequence and a HindIII restriction site was added at the 3' end of the gene. The gene sequence (SEQ ID NO.5) was synthesized and cloned into the Escherichia coli expression vector pET-28a to obtain a recombinant expression vector pET-28a-NMC (SEQ ID NO.6), which contained a strong T7 promoter , lca lactose operon, kanamycin re...
Embodiment 3
[0109] Example 3: Spinning spider silk in pH 4 coagulation bath
[0110] 1. Construction of recombinant expression vectors and strains
[0111] ①Construction of recombinant expression vector
[0112] The N-terminal domain (SEQ ID NO.1) of E.australis spider MaSp1 protein, the C-terminal domain (SEQ ID NO.2) of A.ventricosus spider and the core domain (SEQ ID NO.3) of C.moluccensis origin ) is fused by genetic engineering technology, and codon optimization is performed on the gene according to the expression preference of Escherichia coli without changing its amino acid sequence (SEQ ID NO.4). A NdeI restriction site was added at the 5' end of the gene sequence and a HindIII restriction site was added at the 3' end of the gene. The gene sequence (SEQ ID NO.5) was synthesized and cloned into the Escherichia coli expression vector pET-28a to obtain a recombinant expression vector pET-28a-NMC (SEQ ID NO.6), which contained a strong T7 promoter , lca lactose operon, kanamycin re...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
| Tensile strength | aaaaa | aaaaa |
| Toughness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com