Cloning and transcription carrier construction of cymbidium mosaic virus strain
A technology for building orchid leaves and virus strains, applied in the field of molecular biology, can solve the problems of inability to obtain virus sequences, inability to obtain subgenomes, and ignoring the existence of quasi-species of virus strains
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Embodiment 1-Determination of the sequence of the orchid mosaic virus strain
[0051] This embodiment is to establish the determination of the orchid mosaic virus strain sequence, which includes the following aspects:
[0052] (1) Processing of high-throughput data;
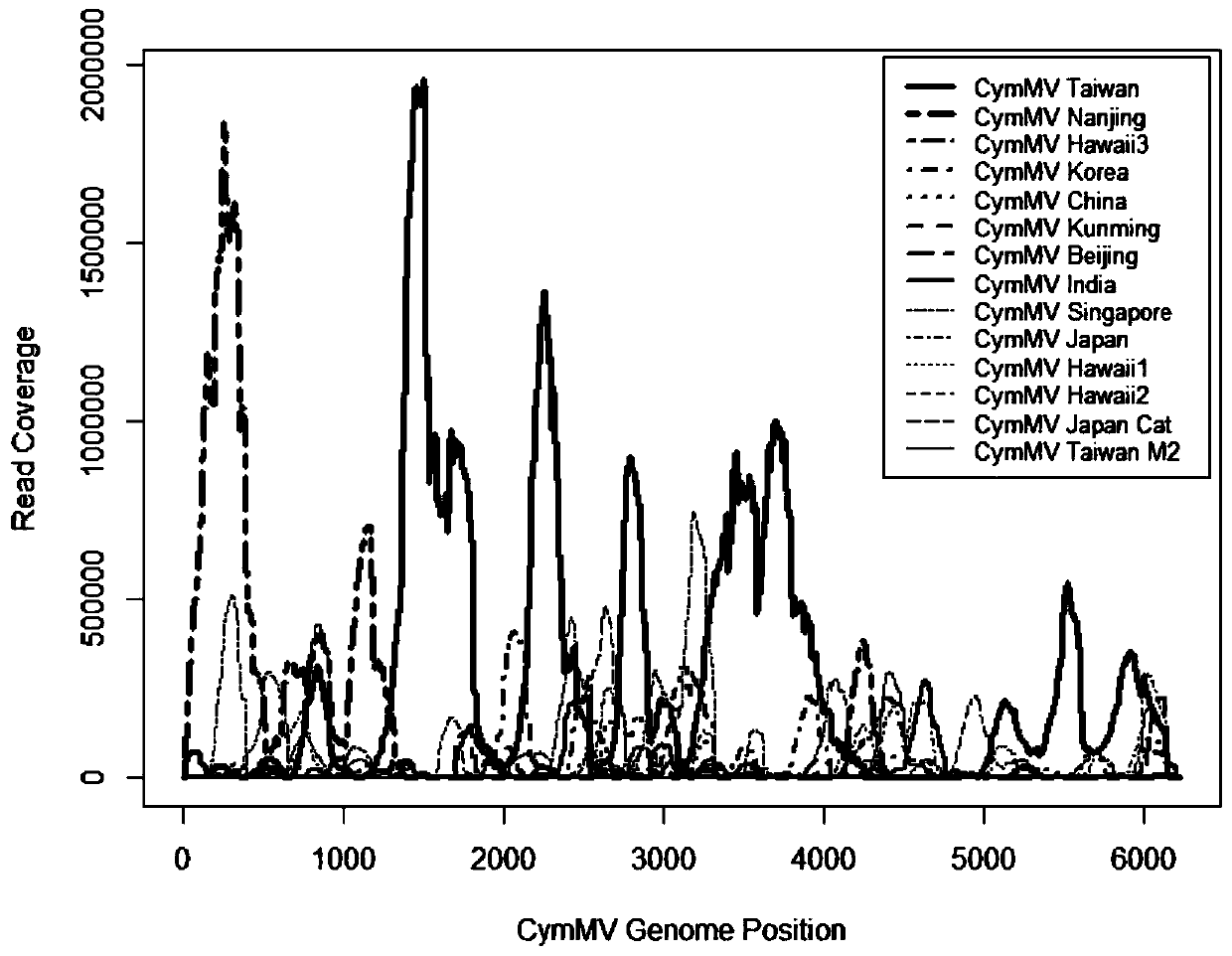
[0053] Using Bowetie2 to compare the high-throughput data to 14 known full-length genome sequences of Jianlan mosaic virus, and using genomeCoverageBed to calculate the read coverage (ReadCoverage) of each position in the Jianlan mosaic virus genome. The 14 sequences were aligned at the alignment position, and the Read Coverage of the position where the sequence was missing was recorded as 0. Use the Plot function in R to make graphs, see figure 1 .
[0054] Each sequence is abbreviated as CymMV, Cymbidium mosaic virus—Taiwan[AY571289.1]; Nanjing[JQ860108.1]; Korea[AF016914.1]; China[KR185347.1]; Kunming [AM055640.2]; ]; India[AM055720.1]; Singapore[U62963.1]; Japan[AB197937.1]; TaiwanM2[EU314803.1]; J...
Embodiment 2
[0071] Construction of embodiment 2-infectious cDNA vector
[0072] This embodiment is the construction of the in vitro transcription vector of the full-length genome sequence of CymMV and the 5' end subgenome of CymMV in Example 1, which adopts the following primers:
[0073] The sequence of the forward primer introduced into the T7 promoter is:
[0074] aaaacgacggccagtgaattcATAATACGACTCACTATAGGGAGAGGAA (SEQ ID NO: 22)
[0075] The reverse primer introduces the PolyA site and the SmaI blunt end restriction site:
[0076] gcctctgcagtcgacggggcccACCCGGGTTTTTTTTTTTTTTTT (SEQ ID NO: 23)
[0077] The full-length and subgenome fragments of CymMV were respectively connected to the vector pUC57, and the sequences of the in vitro transcription vectors of the full-length genome sequence of CymMV and the 5' end subgenome of CymMV were respectively shown in SEQ ID NO: 24 and SEQ ID NO: 25, Its structural diagram is as image 3 As shown, the constructed in vitro transcription vector co...
Embodiment 3
[0078] Preparation and infection verification of embodiment 3-CymMV virus RNA
[0079] This embodiment uses the in vitro transcription vector constructed in Example 2 to carry out the preparation and infection verification of CymMV virus RNA. The specific steps include:
[0080] (1) Linearization of in vitro transcription vectors;
[0081] After the vector was digested with restriction endonuclease SmaI, it was detected by agarose gel electrophoresis to ensure that the vector was completely linearized.
[0082] (2) Synthesis of in vitro transcripts;
[0083] Add 2μL of 10× in vitro transcription buffer, 2μL of 100mM ATP, 2μL of 100mM CTP, 2μL of 100mM UTP, 2μL of 20mM GTP, 4μL of 40mM m7G(5')ppp(5')G, and 4μL of linearized template in a 1.5mL EP tube , T7 RNA polymerase (NEB company) 2uL, 37 ° C water bath for 2 hours, agarose electrophoresis to detect the quality and concentration of transcripts.
[0084] (3) Inoculation of in vitro transcripts;
[0085] The transcribed R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com