Method for detecting BCR-ABL fused gene ABL kinase domain mutation
A technology that fuses genes and kinase regions, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 COLD-PCR combined with next-generation sequencing method to detect mutations in the ABL kinase region of the BCR / ABL fusion gene in peripheral blood
[0033] COLD-PCR combined with next-generation sequencing method to detect BCR-ABL fusion gene in peripheral blood Simultaneous detection of ABL kinase region drug resistance mutation detection kit in two types of M-bcr and m-bcr, including: RNA extraction solution: Red blood cell lysate, Trizol, chloroform, absolute ethanol; reverse transcription reagent: SuperScript II Reverse Transcriptase (ThermoFisher Scientific); detection system PCR reaction solution; library construction reaction solution; Miseq platform sequencing reaction solution; positive control, negative control and blank Control substance.
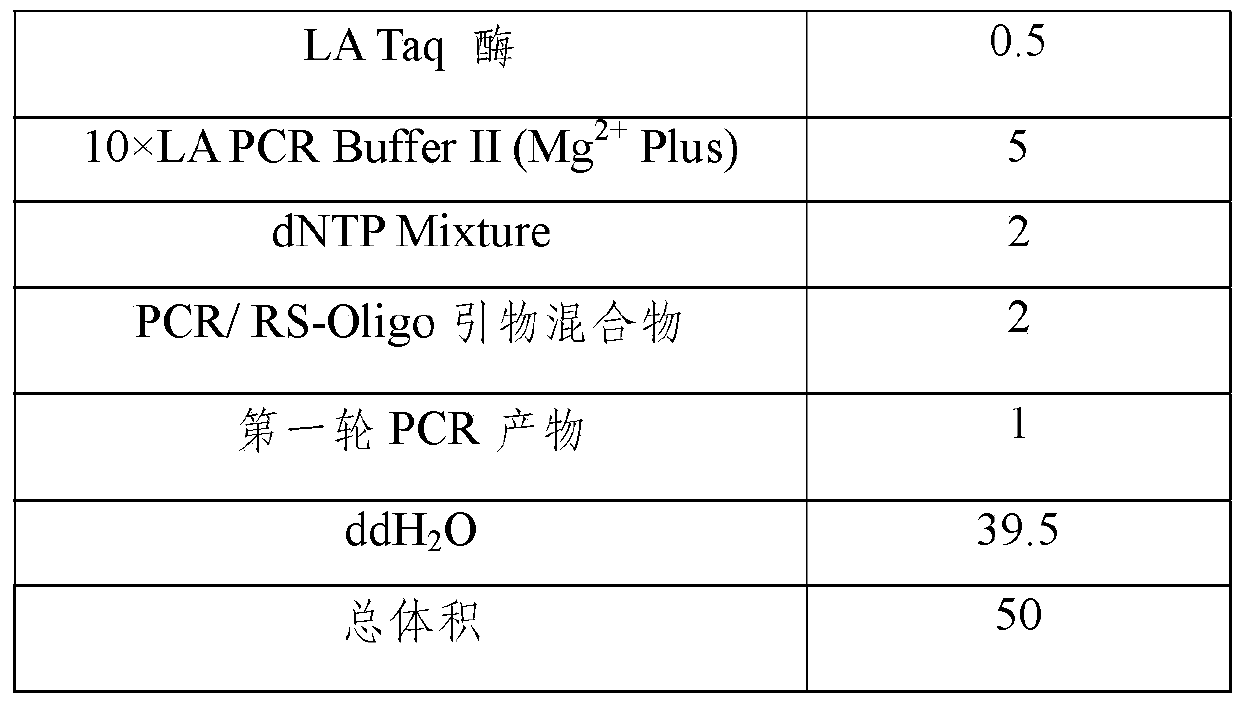
[0034] Detection system PCR reaction solution includes: 10×LA PCR Buffer II (Mg 2+Plus); dNTP Mixture (10mMeach); LA Taq enzyme; primers for amplifying BCR / ABL fusion gene M-bcr and m-bcr spliced BCR-ABL fus...
Embodiment 2
[0081] Example 2 COLD-PCR combined with next-generation sequencing method to detect mutations in the ABL kinase region of the BCR / ABL fusion gene in bone marrow
[0082] COLD-PCR combined with next-generation sequencing method to detect BCR-ABL fusion gene in bone marrow Simultaneous detection of ABL kinase region drug resistance mutation detection kit in two shear types M-bcr and m-bcr, including: RNA extraction solution: red blood cells Lysis solution, Trizol, chloroform, absolute ethanol; reverse transcription reagent: SuperScript II Reverse Transcriptase (ThermoFisher Scientific); detection system PCR reaction solution; library construction reaction solution; Miseq platform sequencing reaction solution; positive control, negative control and blank Control substance.
[0083] Detection system PCR reaction solution includes: 10×LA PCR Buffer II (Mg 2+ Plus); dNTP Mixture (10mMeach); LATaq enzyme; amplification of BCR / ABL fusion gene M-bcr and m-bcr spliced BCR-ABL fusion ...
Embodiment 3
[0130] Example 3 Comparison of the method of COLD-PCR combined with next-generation sequencing and the method of traditional PCR+next-generation sequencing to detect mutations in the ABL kinase region of the BCR-ABL fusion gene
[0131] 1. T-A cloning method to construct the T315I mutant plasmid, the steps are as follows:
[0132] 1.1 Take bone marrow samples from CML patients with T315I mutation, extract RNA, and reverse transcribe cDNA;
[0133] 1.2 Using cDNA as a template and using the sequences shown in SEQ ID NO.1 and SEQ ID NO.3 as primers, amplify the M-bcr type BCR-ABL fusion gene;
[0134] 1.2.1 BCR-ABL fusion gene amplification reaction system:
[0135] Table 21
[0136] Element
Amount added (μl)
LATaq enzyme
0.2
10×LAPCR Buffer II (Mg 2+ Plus)
2
dNTP Mixture
0.8
primer mix
2
cDNA
5
wxya 2 o
10
total capacity
20
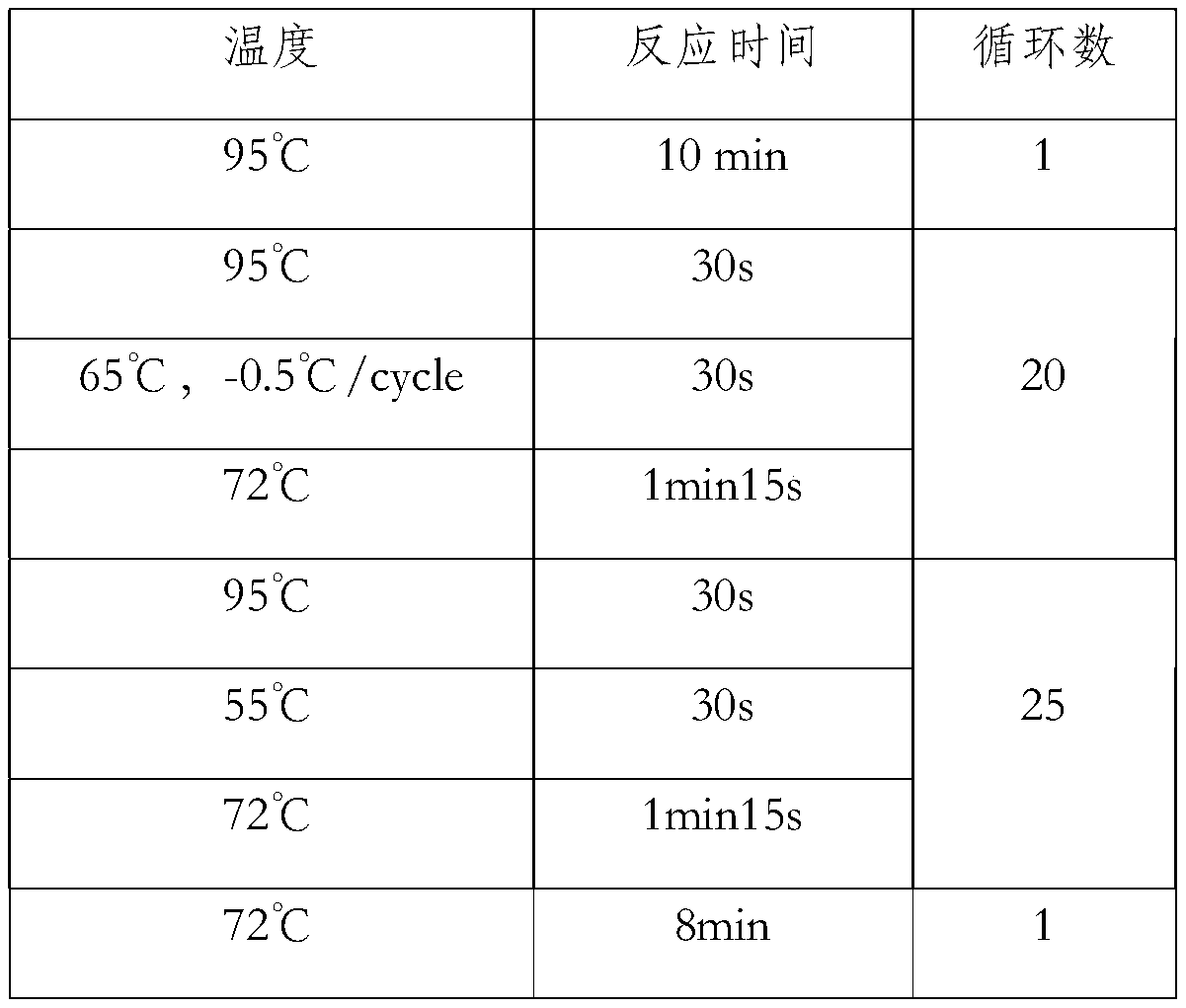
[0137] 1.2.2 BCR-ABL fusion gene amplification procedure: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com