New method for gene point mutation repair
A point mutation and new method technology, applied in the field of gene repair, can solve the problems of incomplete meiosis of germ cells, Msh5 can not run normally, germ cell death and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1 The construction of vector
[0037] 1. Design of sgRNA: Use the sgRNA online design tool to select a suitable sgRNA near the mutation site.
[0038] sgRNA selects Guide#1 and Guide#8, and the sequence information is:
[0039] Guide #1 CTATCGTAGCGCCCGGACCAAGG
[0040] Guide #8 CAAGGAGCTGTACACGCTGCTGG.
[0041] 2. Construct the sgRNA onto the pCS (eGFP) vector, which carries the Cas9 protein.
[0042] (1) Design primers: introduce restriction site BbsI into the upstream and downstream primers, and the primer sequences are:
[0043] Guide #1: F-CACCGCTATCGTAGCGCCCGGACCA
[0044] R-AAACTGGTCCGGGCGCTACGATAGC
[0045] Guide #8: F-CACCGCAAGGAGCTGTACACGCTGC
[0046] R-AAACGCAGCGTGTACAGCTCCTTGC
[0047] (2) Primers anneal into double strands: primers are annealed after being dissolved in TE to 200 μM
[0048] Annealing system total 20μL
[0049]
[0050] After mixing, put it directly into boiling water and anneal until it cools to room temperature, and p...
Embodiment 2
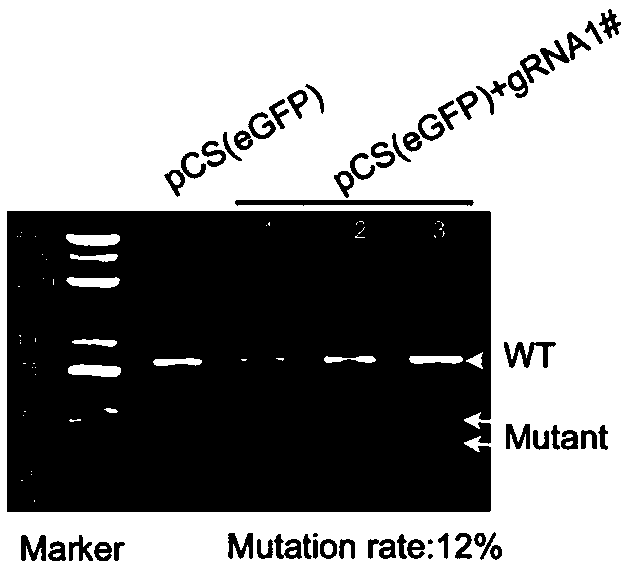
[0061] Example 2 Detection of Cutting Efficiency of Vector
[0062] 1. Guide#1 Cutting Efficiency Detection
[0063] The constructed vector was extracted with an endotoxin-free plasmid extraction kit.
[0064] 1. Pronuclear injection of fertilized eggs, identification of blastocysts, and verification of cleavage activity
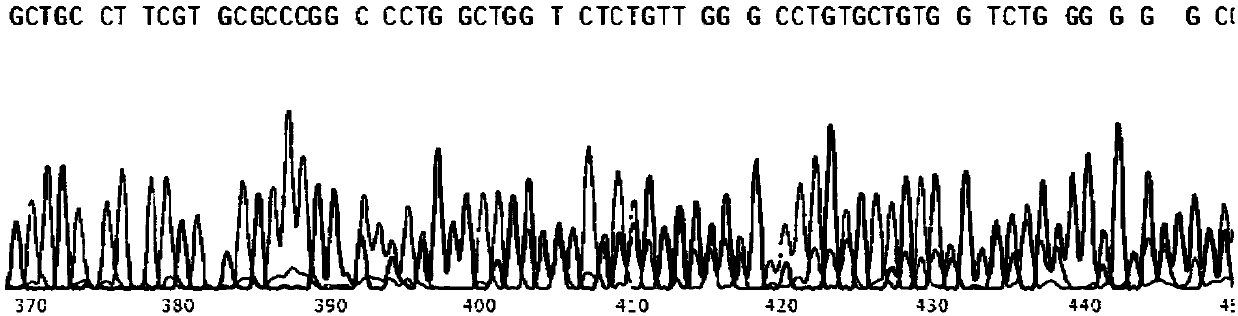
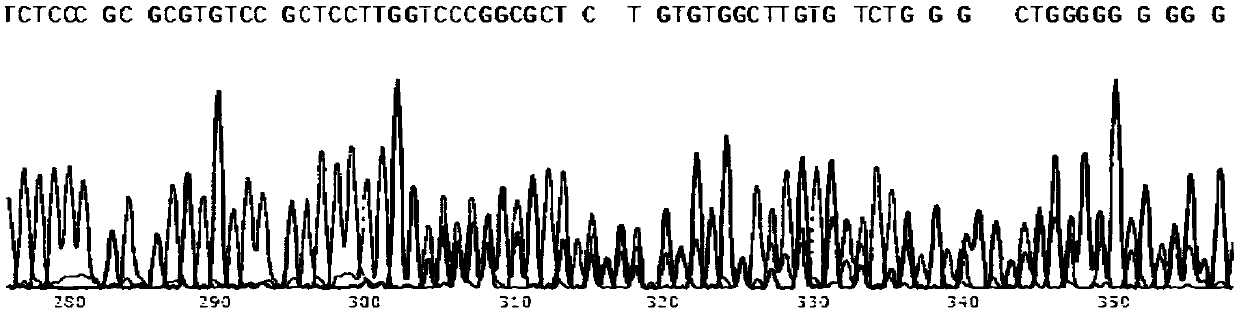
[0065] The carrier is injected into the pronuclear fertilized egg, and the fertilized egg is developed to the blastocyst stage (about 4 days), and the blastocyst is identified to detect whether the cutting is correct. Blastocysts were lysed overnight in a water bath at 56°C by adding 99 μL of lysis solution and 1 μL of proteinase K. The cleaved product was treated at 95°C for 5 minutes to denature proteinase K, and the product could be used for PCR. Blastocyst identification uses Extag for PCR amplification, and the PCR products of the target bands are sent for sequencing.
[0066] Extag PCR system 50μL
[0067]
[0068] Extag PCR reaction program
[...
Embodiment 3
[0098] Example 3 Design Doner DNA
[0099] Select a correct sequence near the upstream and downstream of the mutation site as Donor, and send it to Synthetic. The sequence is:
[0100] CAGTTTCTCTCAGAGGACAAGCTGCACTATCGTAGCGCCCGGACCAAGGAGCTGGACACGCTGCTGGGAGACCTGCACTGCGAGATCCGGGGTGAGGAGCCCGTGGTAGGAGGGGGCAGGCTGCTCTAAC.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com