EST-SSR marker development and application of rhododendron pulchrum sweet
A rhododendron and splendid technology, applied in the field of SSR molecular marker development and cross-species amplification application, can solve the problems of limitation and limited microsatellite markers, and achieve the effect of high versatility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
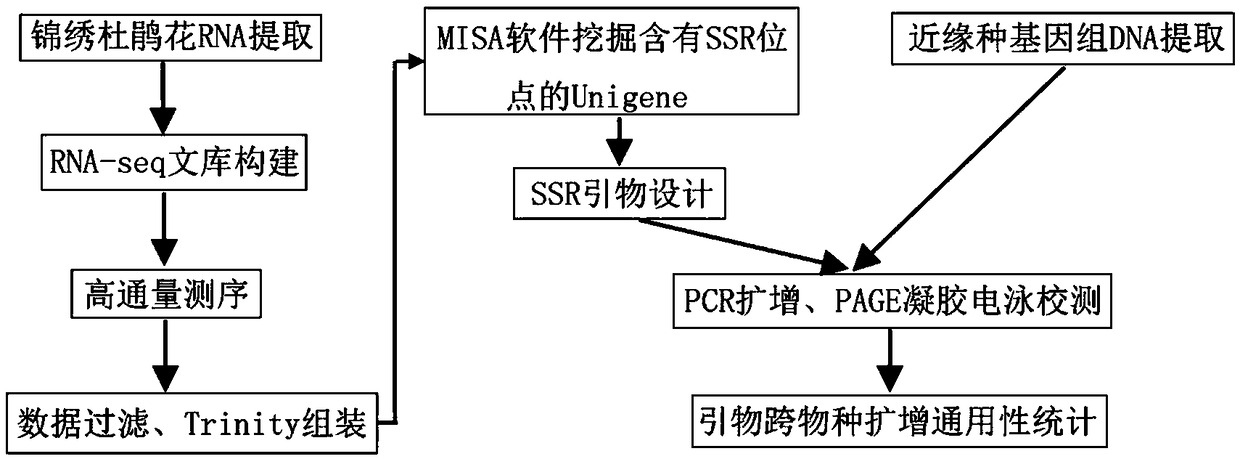
[0040] [Example 1] Development of EST-SSR primers for Rhododendron splendens with high polymorphism
[0041] Based on the high-throughput sequencing of transcriptome provided by the present invention, combined with bioinformatics methods for SSR sequence search and SSR marker primer design and verification, the specific implementation methods are as follows:
[0042](1) Screening of Unigene sequences rich in SSR sites: Total RNA was extracted from Rhododendron splendens, a transcriptome sequencing library was constructed, and high-throughput sequencing was performed using Illumina next-generation sequencer. The sequencing data was assembled after strict filtering to obtain Unigene, which was used as background data for subsequent SSR primer development. Use MISA software to search for SSR sites in Unigene. The search criteria are: dinucleotides, trinucleotides, tetranucleotides, pentanucleotides, and hexanucleotides. The minimum repeat times are 10, 8, 6, respectively. 5, 4 t...
Embodiment 2
[0048] [Example 2] Application of 15 pairs of EST-SSR primers in the study of genetic diversity of Rhododendron splendens
[0049] (1) DNA extraction: A total of 35 individuals of Rhododendron splendens were collected in the Dabie Mountains of Hubei Province (the collection location is distributed at 114.56°-115.043°E east longitude, 31.03°-31.68°N north latitude, and 220-650m above sea level), and the numbers are numbered For JXDJ1-JXDJ30. The genomic DNA of the leaf tissue was extracted by 3×CTAB method, and the quality was detected by 1% agarose gel electrophoresis and the concentration was determined by ultraviolet spectrophotometry. After dilution to 100ng / μl, it was stored in a -20°C refrigerator for later use.
[0050] (2) PCR amplification: 20μl reaction system contains: ddH 2 O 15.7μl, 10×Buffer (containing Mg 2+ ) 1.5 μl, dNTPs (10 mM) 0.2 μl, 10 μM forward and reverse primers 0.2 μl each, Taq DNA polymerase 0.2 μl, DNA template (50ng / μl) 1 μl. The reaction progra...
Embodiment 3
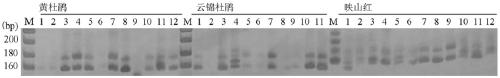
[0056] [Example 3] Detection of 15 pairs of EST-SSR primers for cross-species amplification in 3 related species of Rhododendron splendens
[0057] The general detection steps of EST-SSR primers in three Rhododendron splendens relatives:
[0058] (1) Genomic DNA extraction: 3 × CTAB method was used to extract genomic DNA from fresh leaf tissue of three Rhododendron splendens relatives: liquid nitrogen grinding, phenol / chloroform (volume ratio 1:1) extraction, isopropanol precipitation , 70% ethanol washing, and RNase treatment, the extracted DNA was dissolved in 50 μL TE solution.
[0059] (2) PCR amplification. Use a 15 μl reaction system containing ddH 2 O 11.6μl, 10×Buffer (containing Mg 2+ ) 1.5 μl, dNTPs (10 mM) 0.2 μl, forward and reverse primers (10 μM) 0.2 μl each, Taq DNA polymerase (2.5U / μl) 0.3 μl, DNA template (50ng / μl) 1 μl. The reaction program was set as follows: pre-denaturation at 94°C for 5 min, 35 amplification cycles (denaturation at 94°C for 40 sec, am...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com