A seedless tomato with regular cracks and bright color and its cultivation method
A technology with bright colors and cracks, applied in the field of plant genetic engineering, can solve the problems of single gene function loss, homologous gene function compensation failure, high coding sequence homology, gene function redundancy, etc., to achieve low cost and future generations inheritance Simple, Vigorous Effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
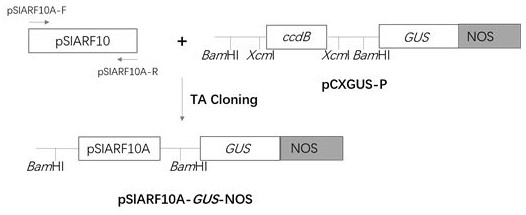
[0031] Example 1 Tomato Gene SlARF10A Cloning of the promoter and analysis of GUS histochemical staining
[0032] (1) SlARF10A Cloning of gene promoters
[0033] Using the CTAB method (RTG2405-01, Zhongke Tairui) to extract tomato genomic DNA, according to the sequence shown in SEQ ID NO.3 pSlARF10A Nucleotide sequence, design primers pSlARF10A -F and pSlARF10A -R uses the extracted genomic DNA as a template, with pSlARF10A -F (forward primer) and pSlARF10A -R (reverse primer) is the primer for the gene SlARF10A expansion.
[0034] The primer sequences are as follows:
[0035] pSlARF10A -F:5'-CAATTTACATATACAACACATGCCTCA-3'
[0036] pSlARF10A -R:5'-GCTAATCCAATAGTTTTTCCCCTTC-3'
[0037] P CR amplification system: high-fidelity amplification enzyme Pprime STAR HS (R010A, TaKaRa) 0.25 μL, 5× P primeSTAR Buffer (Mg 2+ P lus) 5 μL, forward primer (10 μM) 0.5 μL, reverse primer (10 μM) 0.5 μL, template (DNA) 1 μL, dNT P (2.5mM) 2μL, sterile ddH 2 O to m...
Embodiment 2
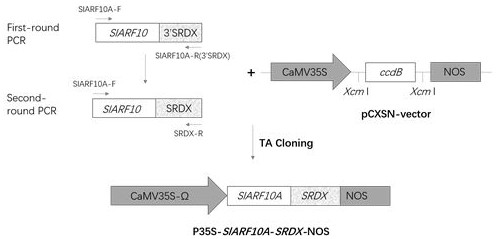
[0059] Example 2 tomato SlARF10A-SRDX Fusion Gene Cloning and Genetic Transformation Mediated by Agrobacterium
[0060] (1) SlARF10A-SRDX Cloning of fusion fragments
[0061] Take Micro Tom tomato mixed samples (leaves, flowers and fruits) as materials, use TRIzol™ Plus RNAPurification Kit (12183555, Invitrogen™), extract tomato total RNA according to the instructions, and use DNaseI (18047019, Invitrogen™) to remove residual trace DNA, And use a spectrophotometer to measure the concentration of RNA for later use.
[0062] Take about 2.0 μg tomato total RNA, using P crimeScri p t II first-strand cDNA synthesis kit (6210A, Takara), and synthesize the first strand of cDNA according to the instructions.
[0063] According to the tomato shown in SEQ ID NO.1 SlARF10A Gene sequence, design of specific primers SlARF10A -F, SlARF10A- 3'SRDX- R (without the termination password) and SRDX -R. Using cDNA as a template, with SlARF10A -F (forward primer) and SlARF10A - ...
Embodiment 3
[0102] Example 3 Tomato SlARF10B-SRDX Cloning of Fusion Gene and Genetic Transformation Mediated by Agrobacterium
[0103] (1) SlARF10B-SRDX Cloning of fusion fragments
[0104] Take Micro Tom tomato mixed samples (leaves, flowers and fruits) as materials, use TRIzol™ Plus RNAPurification Kit (12183555, Invitrogen™), extract tomato total RNA according to the instructions, and use DNaseI (18047019, Invitrogen™) to remove residual trace DNA, And use a spectrophotometer to measure the concentration of RNA for later use.
[0105] Take about 2.0 μg tomato total RNA, using P crimeScri p t II first-strand cDNA synthesis kit (6210A, Takara), and synthesize the first strand of cDNA according to the instructions.
[0106] According to the tomato shown in SEQ ID NO.3 SlARF10B Gene sequence, design of specific primers SlARF10B -F, SlARF10B- 3'SRDX- R (without the termination password) and SRDX -R. Using cDNA as a template, with SlARF10B -F (forward primer) and SlARF10B...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com