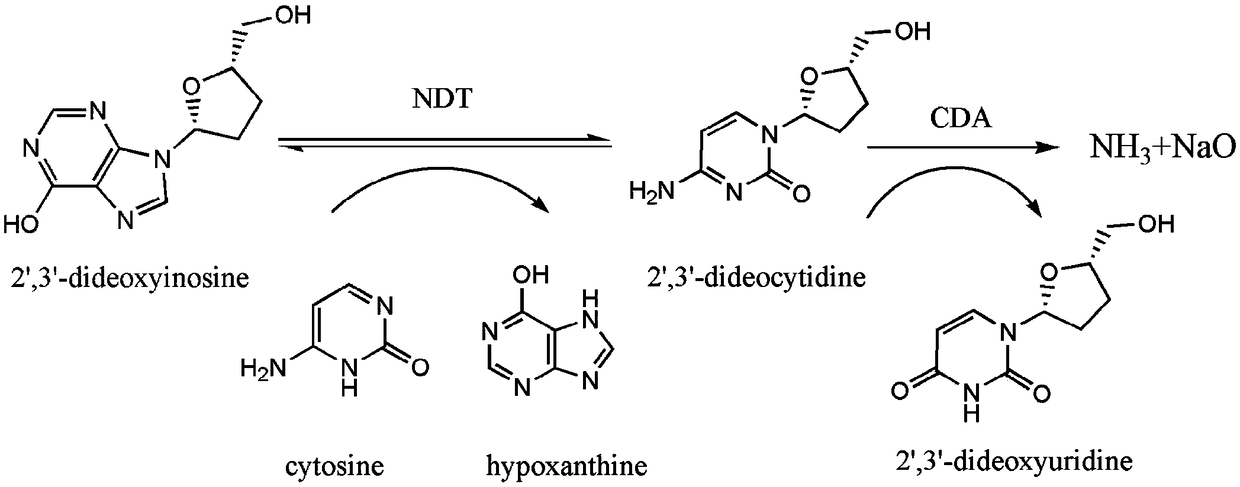
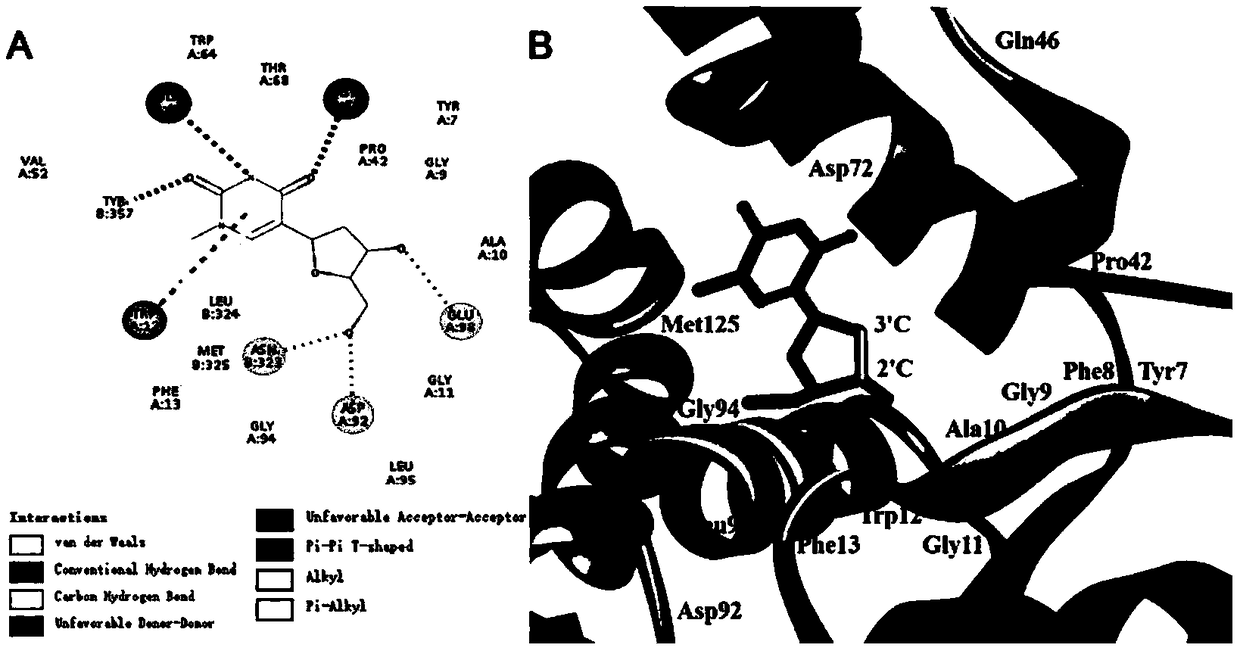
Directed evolution and biocatalytic application of N-deoxyribosyltransferase II
A deoxyribose and transferase technology, applied in the direction of glycosyltransferase, transferase, biochemical equipment and methods, etc., can solve the problem of low catalytic efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1, the acquisition of N-deoxyribosyltransferase II (NDT) mutant
[0047] 1. Construction of N-deoxyribosyltransferase Ⅱ (NDT) recombinant engineering strain
[0048] Lactobacillus helveticus (CGMCC 1.1877) bacterial solution was used as a template to perform PCR amplification to obtain a fragment of the target gene ndt (the nucleotide sequence is sequence 1 in the sequence listing), and the amino acid sequence of the protein encoded by the gene is sequence 2 in the sequence listing.
[0049] According to the nucleic acid sequence of Lactobacillus helveticus N-deoxyribosyltransferase II gene (ndt) found in Genbank, a pair of amplification primers were designed:
[0050] ndt(+):5'–CGCCATATGATGAACAAGAAAAAAGAC-3'
[0051] ndt(-):5'–CGGGAATTCTTAATATACAGCTCCG-3'
[0052] The recovered and purified PCR product was ligated with the pMD18-T plasmid to obtain the cloning plasmid pMD18-T-ndt. The ligation product pMD18-T-ndt was transformed into Escherichia coli Trans5...
Embodiment 2
[0068] Example 2, Preparation and functional testing of NDT mutant THNDT
[0069] 1. Preparation of NDT mutant THNDT
[0070] 1. Recombinant bacteria expressing NDT mutant THNDT
[0071] The recombinant bacterium expressing the NDT mutant THNDT is a recombinant bacterium obtained by introducing the recombinant plasmid pET-28a(+)-THNDT into the expression strain E.coli BL21(DE3). The above-mentioned recombinant plasmid pET-28a(+)-THNDT is obtained by replacing the fragment between the Nde I and EcoR I restriction sites of the pET-28a(+) vector with the NDT mutant THNDT coding gene shown in Sequence 1 in the sequence listing A plasmid expressing the NDT mutant THNDT.
[0072] The recombinant bacterium expressing NDT is the recombinant bacterium obtained by introducing the recombinant plasmid pET-28a(+)-NDT into the expression strain E.coli BL21(DE3). The above-mentioned recombinant plasmid pET-28a(+)-NDT is a plasmid obtained by replacing the fragment between the Nde I and Ec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com