Corbicula fluminea mitochondrial genome enrichment, extraction and indentifying method
A technology of mitochondrial genome and identification method, applied in the field of molecular biology, can solve the problems of difficult to achieve, low total amount, low extraction purity, etc., and achieves the effect of more total amount, good discrimination, and good DNA integrity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
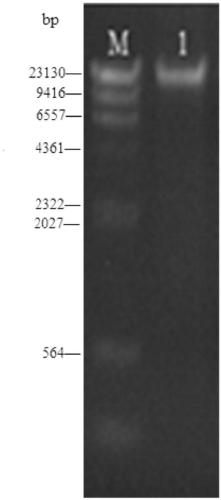
[0044] Example 1 Extraction of Mitochondrial Genomic DNA from River Clams by Differential Centrifugation
[0045] Take 5g of fresh river clam collected in Jiangsu, China, take its muscle and grind it thoroughly in a pre-cooled glass homogenizer, filter the ground sample with a 1um filter membrane, and centrifuge the collected filtrate with a centrifugal force of 3000g 20 minutes. Transfer the centrifuged supernatant to a new centrifuge tube, centrifuge at 15,000g for 20 minutes, collect the precipitate, add 1ml suspension buffer to the precipitate, and gently suspend the precipitate with a brush. Centrifuge the suspension at 3000g for 10 minutes, take the supernatant, add 2ug DNase I per gram of fresh weight, digest at 35°C for 2 hours, add 0.5mol / L EDTA to stop the digestion reaction, centrifuge at 18000g for 20 minutes, collect the precipitate and get the crude Lift the mitochondria.
[0046] Add 1 / 10 volume of 10% SDS and 1 / 100 volume of 30mg / ml RNase to the crudely extra...
Embodiment 2
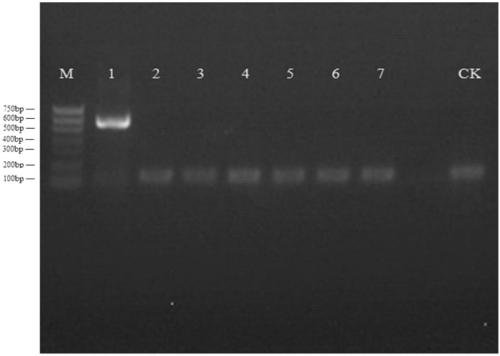
[0061] Embodiment 2 primer pair Mit1 amplification and identification
[0062] This study is divided into river clam group, Cyprinidae: silver globber group, Bayesian group, wheat ear fish group, similar bream group, triangular sail mussel group and patina group.
[0063] The operation method of mitochondrial genomic DNA enrichment and extraction of samples to be identified is the same as that in Example 1.
[0064] Using the mitochondrial genomic DNA of each sample to be identified as a template, the primer pair Mit1:Mit1F:5'-ATGGTTTAACGGCTGCGATTGAA-3'(SEQ ID No.3) and Mit1R:5'- CCAACATCGAGGTAGCAAACTTTCT-3' (SEQ ID No. 4) was PCR amplified.
[0065] Before performing PCR amplification, the reagents used were put on ice for thawing, the concentrations of the primers were diluted to 5umol / L, and the total PCR reaction system was 20ul. The reaction system is as follows: 5×FastPfu Buffer 4ul, 2ul 2.5mMdNTPs, Forward Primer (5μM) and Reverse Primer (5μM) 0.8ul each, FastPfu Po...
Embodiment 3
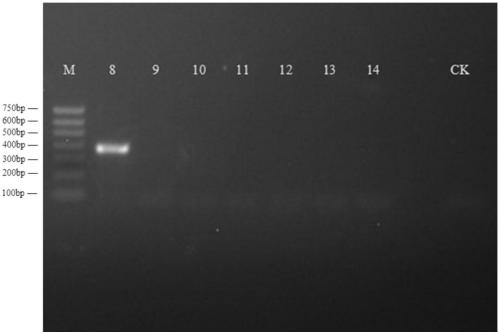
[0073] Embodiment 3 primer pair Mit2 amplification and identification
[0074] This study is divided into river clam group, Cyprinidae: silver globber group, Bayesian group, wheat ear fish group, similar bream group, triangular sail mussel group and patina group.
[0075] The operation method of mitochondrial genomic DNA enrichment and extraction of samples to be identified is the same as that in Example 1.
[0076] Using the mitochondrial genomic DNA of each sample to be identified as a template, the primer pair Mit2: Mit2F:5'-ATCTTACGGGTACTACGAACAAACG-3'(SEQ ID No.5) and Mit2R:5'-ACACCTCTACGGACGCCTCT- 3' (SEQ ID No.6) was amplified by PCR.
[0077] Before performing PCR amplification, the reagents used were put on ice for thawing, the concentrations of the primers were diluted to 5umol / L, and the total PCR reaction system was 20ul. The reaction system is as follows: 5×FastPfu Buffer 4ul, 2ul 2.5mMdNTPs, Forward Primer (5μM) and Reverse Primer (5μM) 0.8ul each, FastPfu Poly...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com