Application of snp site of clip3 gene
A site, aortic dissection technology, applied in the field of biomedical detection, can solve the problems of difficult early risk assessment, difficult early detection, high risk of AD blood vessels, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
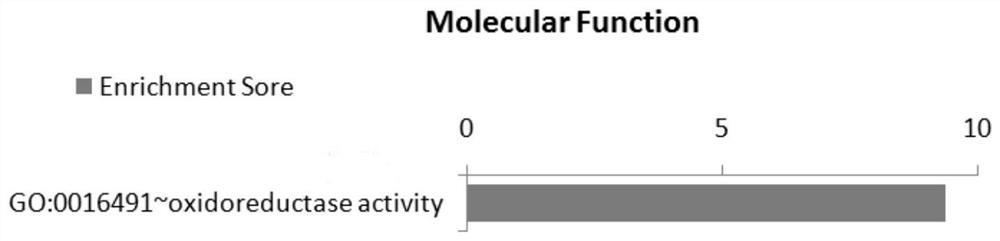
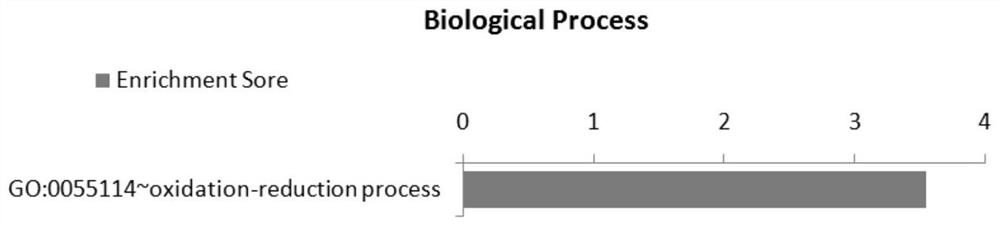
Image
Examples
Embodiment 1
[0029] Example 1 Sample Collection
[0030] From January 2017 to April 2018, 59 patients with Stanford type A aortic dissection were diagnosed by aortic CTA in Shenzhen Sun Yat-sen Cardiovascular Hospital, and 2 mL of whole blood and 590 normal control samples were collected from the searched by Makino Sequencing Company. database. Informed consent was obtained from the patient and reviewed by the Ethics Committee.
[0031] Sample processing: EDTA anticoagulated whole blood was mixed with Trizol at a ratio of 1:1, mixed thoroughly and placed in a 1.8mL cell cryopreservation tube, cooled rapidly in liquid nitrogen for 30 seconds and stored in a -80°C refrigerator.
Embodiment 2
[0032] Embodiment 2 extracts blood sample DNA
[0033] (1) Add 1 mL of CL cell lysate to 1 mL of EDTA (0.01M, China, Huamei Bioengineer) anticoagulated blood, gently invert and mix 6 times, centrifuge at 3600 rpm for 5 min, and discard the supernatant;
[0034] (2) Pour 1mL of CL cell lysate into the centrifuge tube again, gently invert and mix 6 times, centrifuge at 3600rpm for 5min, discard the supernatant; under the premise of ensuring that the precipitate remains in the tube, invert the centrifuge tube Stand on clean absorbent paper for 2 minutes;
[0035] (3) configuring a mixed solution of proteinase K and buffer FG;
[0036] (4) Add 500 μL of proteinase K and buffer FG mixture, then mix until the solution has no lumps;
[0037] (5) Water bath at 65°C for 30 minutes, and mix by inverting several times during the period;
[0038] (6) Add 1 mL of isopropanol, then invert and mix until clustered or filamentous genomic DNA appears;
[0039] (7) Centrifuge at a speed of 3...
Embodiment 3
[0047] Example 3 Whole Exome Sequencing
[0048] 1. DNA library construction
[0049] 3 μg of DNA was ultrasonically fragmented, the ends of the interrupted fragments were blunted, A was added to the 3' end, and the adapter was connected, and the fragments between 350 and 400 bp were selected to prepare the genome-wide library. Library samples were quality controlled using an Agilent 2100 bioanalyzer (Agilent Technologies, USA).
[0050] 2. Target region capture sequencing
[0051] Whole-exome detection was performed using GenCap liquid-phase capture target gene technology (Beijing Mikino Company). Mix 1 μg of DNA library with BL buffer and probe, heat at 95°C for 7 minutes, heat at 65°C for 2 minutes, add 23 μL of HY buffer preheated to 65°C, and hybridize at 65°C for 22 hours. 50 μL MyOne magnetic beads (Life Technology, USA) were washed 3 times with 500 μL 1X binding buffer, and resuspended in 80 μL 1X binding buffer. Add 64 μL 2X Binding Buffer to the hybridization mix...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com