Novel xylanase
A new technology of xylanase, applied in the field of genetic engineering, can solve the problems of restricting large-scale applications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
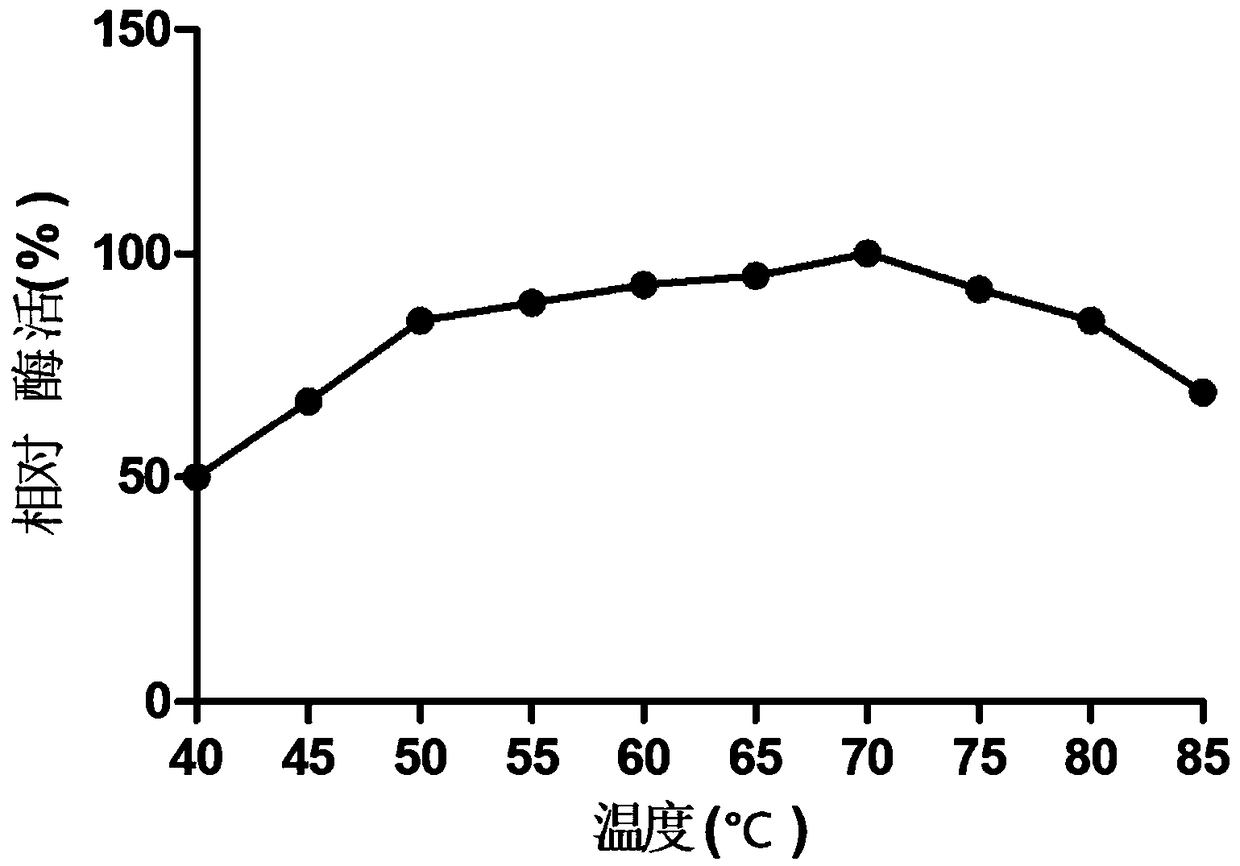
Image
Examples
Embodiment 1
[0021] Embodiment 1, the cloning of Xyn-dx gene and the construction of recombinant plasmid pET-30a / Xyn-dx
[0022] The cloning method of Xyn-dx xylanase gene is as follows:
[0023] ① Design primers containing two restriction enzyme sites of EcoR I and Xho I:
[0024] LXY-F: 5'CCGGAATTCATGCGCCCCGTAGACTAT 3', the nucleotide sequence is shown in SEQ ID No.3,
[0025] LXY-R: 5'CCGCTCGAGGATCATTCTTATTCTAAA 3', the nucleotide sequence is shown in SEQ ID No.4.
[0026] ②PCR amplification of the pre-prepared positive plasmid of the Xyn-dx gene (the Xyn-dx carried on the plasmid is obtained by screening the microbiota construction library in the rumen of the cow), and the amplification conditions of the Xyn-dx gene are as follows: 94 Pre-denaturation at ℃ for 2 minutes; denaturation at 94℃ for 30s, annealing at 58℃ for 30s, extension at 72℃ for 45s, a total of 30 cycles; in the 30th cycle, extension at 72℃ for 10min, PCR products were identified by gel electrophoresis, recovered by ...
Embodiment 2
[0030] Embodiment 2, the expression of recombinant plasmid in escherichia coli
[0031] Heat shock transformation Take 10 μl of the ligation product and mix it carefully with E. coli competent BL21(DE3), and put it in an ice bath for 30 minutes; heat shock it in a water bath at 42°C for 60 seconds, and immediately take it out of the ice bath for 2-3 minutes; add 900 μl of SOC solution preheated at 37°C, 150rpm Incubate at a constant temperature for 1 hour; absorb 50 μl of the transformed bacterial solution, apply it to a screening medium containing kanamycin, and incubate at a constant temperature at 37°C for 15 hours; pick plaques for PCR identification of the bacterial solution, and transfer the positive clones to Expand culture in LB liquid medium containing kanamycin.
Embodiment 3
[0032] Embodiment 3, induction and purification of positive bacterial strains
[0033] ①Induction of positive strains
[0034] Pipette 10 μl of positive bacterial liquid and transfer to 5 mL LB liquid medium containing kanamycin (100 μg / mL), and culture at 37° C., 220 rpm for 16 hours.
[0035] Transfer 5 mL of bacterial liquid to 100 mL of LB liquid medium, and continue culturing at 37° C. and 220 rpm until OD=0.8 (about 8 hours).
[0036] Add 100 μl IPTG (final concentration 1 mmol / L), induce culture at 25° C., 150 rpm for 12 hours at low temperature.
[0037] Transfer the induced bacterial solution to a 50 mL centrifuge tube, centrifuge at 12000 rpm at 4°C for 15 min, discard the supernatant, and add 20 mL of PBS buffer (pH7.4) to fully resuspend the bacterial cells.
[0038] Put it into a beaker filled with ice cubes, and repeat ultrasonic crushing twice (15 min each time, 3 s working time, 5 s intermittent time, No. 6 horn, 195 W crushing power).
[0039] The crushed b...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com