Expansion protein and xylanase fusion protein, coding gene and applications thereof
A fusion protein and xylanase technology, which is applied in the fields of genetic engineering technology and biomass utilization, can solve the problems of increasing product separation difficulty and cost, weakening the application prospect of xylo-oligosaccharide, weak recognition ability and binding ability of hemicellulose, etc. , to achieve the effect of improving specific activity, reducing separation difficulty and cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
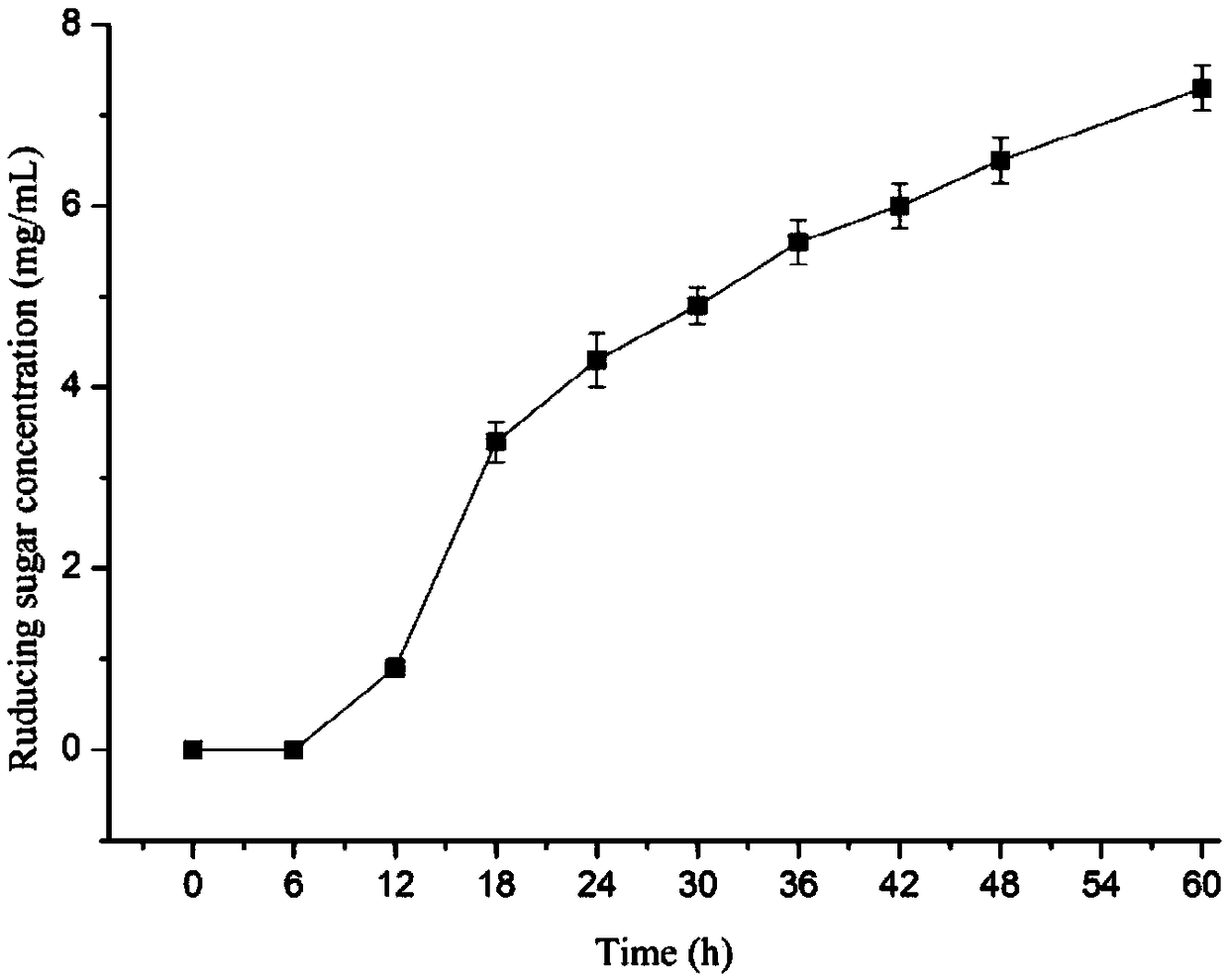
Image
Examples
Embodiment 1
[0024] This example illustrates the acquisition method of the expansin EXCL gene of the present invention.
[0025] This example illustrates the procedure for obtaining the expandin EXCL gene and xylanase XYN gene from Bacillus subtilis LC.
[0026] Bacillus subtilis LC was obtained from the preliminary screening of our laboratory, and the strain preservation registration number is CCTCC No: M208073. . The total DNA of Bacillus subtilis LC genome was extracted by the phenol-chloroform method. According to the analysis of the whole gene sequencing results of Bacillus subtilis 168 (NCBI Reference Sequence: CM000487.1), the expansin EXCL gene and xylanase XYN gene were obtained, and the EXCL and XYN amplification primers were designed according to the gene sequence .
[0027] The EXCLF sequence is: GCCG CCATGGG GCATATGACGACCTGCATG
[0028] The EXCLR sequence is: CCG CTCGAG TTCAGGAAACTGAACATGGC
[0029] The XYNF sequence is: GCCG CCATGGG GCTAGCACAGACTACTGGCA
[0030] T...
Embodiment 2
[0034] This example takes the fusion protein EXCL-EK2-XYN (SEQ ID NO: 7) as an example to illustrate the acquisition of the recombinant fusion protein gene, in which EK2 is a rigid linker peptide (EKKKA) n , n=2.
[0035] Using the Bacillus subtilis LC genome as a template, the genes containing expansin EXCL (SEQ.ID.NO.1) and xylanase XYN (Genbank: KX196201, SEQ.ID.NO.2) were amplified. Design EXCL upstream primers and EXCL downstream primers:
[0036] EXCL upstream primer: GCCG CCATGGGC GCATATGACGACCTGCATG;
[0037] EXCL downstream primers:
[0038] TTTAGCCGCAGCTTCTTTTGCCGCAGCTTCTTCAGGAAACTGAACATGGC.
[0039]The upstream primer introduces the restriction endonuclease NcoI site, and the downstream primer introduces the connecting peptide (EAAAK) 2 coding genes. PCR reaction system 50 μL, in which ddH 2 O 20 μL, 10×prime star buffer mix 25 μL, Bacillus subtilis LC genome 1 μL, EXCLF1 and EXCLR1 (20 μM) each 2 μL. Design XYN upstream primers and XYN downstream primers: ...
Embodiment 3
[0045] This example takes the fusion protein EXCL-EK2-XYN as an example to illustrate the construction of the recombinant expression vector pET-EXCL-EK2-XYN and the preparation of recombinant expression transformants.
[0046] The expansin-xylanase (EXCL-EK2-XYN) PCR product was column-purified and then digested with plasmid pET-28a (+) at 37 °C overnight with NcoI and XhoI, and the purified EXCL-EK2- XYN fragment and linear band of pET-28a (+) vector. Next, use T 4 The two fragments were ligated by DNA ligase, ligated overnight at 16°C, and the ligated product was transformed into Escherichia coli BL21(DE3), spread on LB medium containing 100 μg / mL ampicanamycin and cultured overnight at 37°C. The transformant was verified by colony PCR, and the positive transformant was inoculated into liquid LB medium for expansion culture. The culture medium was extracted with the plasmid mini-extraction kit from TAKARA company and identified repeatedly by NcoI and XhoI double enzyme dig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com