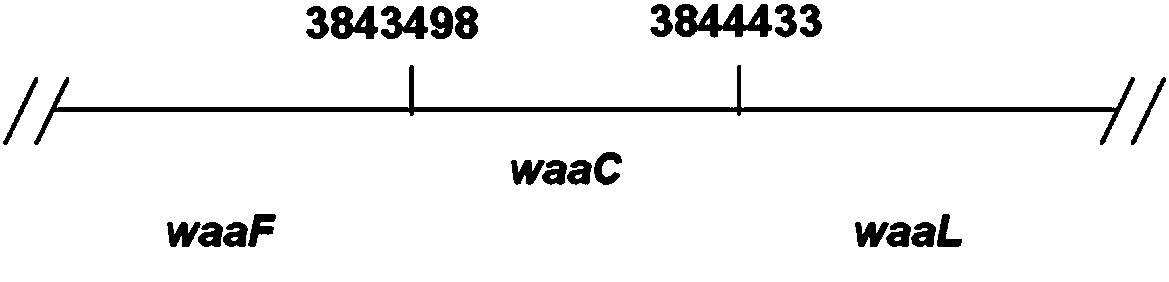Method for evolving host specificity of bacteriophage
A host-specific, bacteriophage technology, applied in the biological field, can solve the problems of lack of understanding of bacteriophage, the challenges of transformation and mass production of unknown bacteriophages, and the impossibility of host-specificity, so as to promote the process of host-specificity and simplify host-specificity. The effect of the sexual process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] The construction of embodiment 1 springboard host
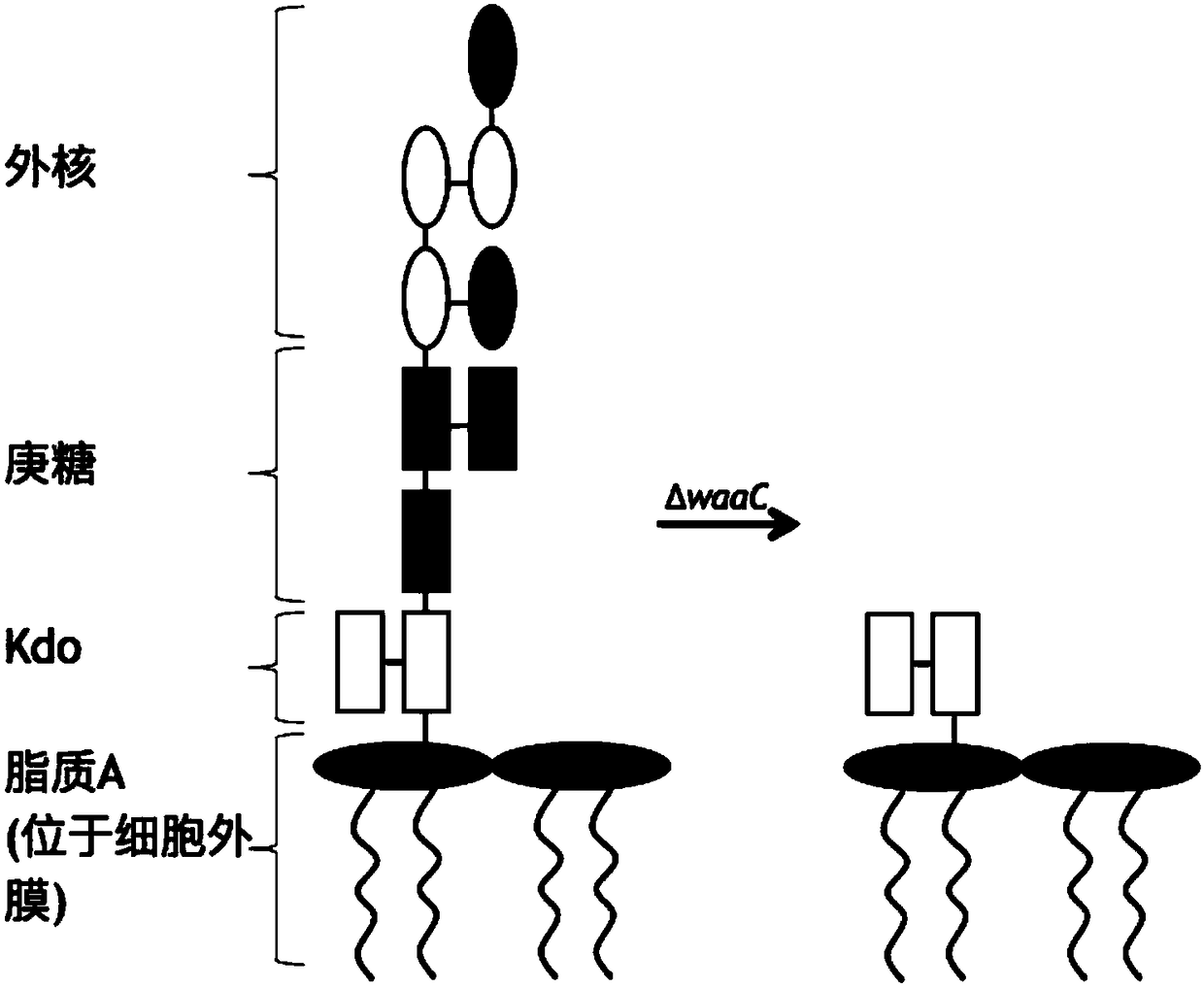
[0047] The Kan resistance gene fragment was amplified by PCR using a pair of primers containing homology arms, and then the corresponding fragment of the waaC gene on the Escherichia coli W3110 genome was replaced by homologous recombination to obtain the correct waaC knockout strain. The primers containing the homology arms are
[0048] F: AGTTTAAAGGATGTTAGCATGTTTTACCTTTATAATGATGATAACTTTTC (SEQ ID NO. 1);
[0049] R: TACTGGAAGAACTCAACGCGCTATTGTTACAAGAGGAAGCCTGACGGATG (SEQ ID NO. 2).
[0050] The PCR reaction system is:
[0051]
[0052] The PCR reaction procedure is:
[0053] 95°C for 5 minutes;
[0054] 28 cycles of 95°C for 30sec, 54°C for 30sec, and 72°C for 1.5min;
[0055] 72°C for 10 minutes.
[0056] The method of homologous recombination is specifically as follows: first, Dpn I is used to treat the PCR product containing the homology arm and the Kan resistance gene fragment, and then the PCR product i...
Embodiment 2
[0062] The construction of embodiment 2 superphage
[0063] Set the plaque forming units (PFU) to 10 7 / ml of T7 phage was shaken and mixed with the springboard host at MOI=1. After standing still for 20 minutes, add LB medium containing 0.6% agar at a temperature of 50°C, shake and mix well, and pour it into the pre-prepared lower layer of LB for solid culture. base (agar content 1.5%), let stand at room temperature for 1 hour until the upper layer of agar is solidified, put it into a 37°C constant temperature incubator, and after overnight cultivation, usually 1-2 phage plaques are produced on a plate, which is "super phage 1.0" .
Embodiment 3
[0064] Example 3 Sequencing of Superphage 1.0
[0065] The products of genes11, 12, and 17 of T7 phage are the main receptor-binding proteins responsible for recognizing and binding LPS, wherein gene11 encodes tail tube protein A, gene12 codes tail tube protein B, and gene17 codes tail fiber protein. Pick the superphage 1.0 obtained in Example 2, use the springboard host to further amplify the culture, then extract the phage DNA, use PCR to amplify T7 phage and gene11, gene12, gene17 fragments of the superphage, perform Sanger sequencing after purification, and then compare Analysis of mutation sites of super phages, the results are as follows image 3 shown.
[0066] Figure 4 The results showed that gene12 carried a base mutation (A was replaced by G), and gene17 carried a base mutation (G was replaced by T), both of which were sense mutations.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com