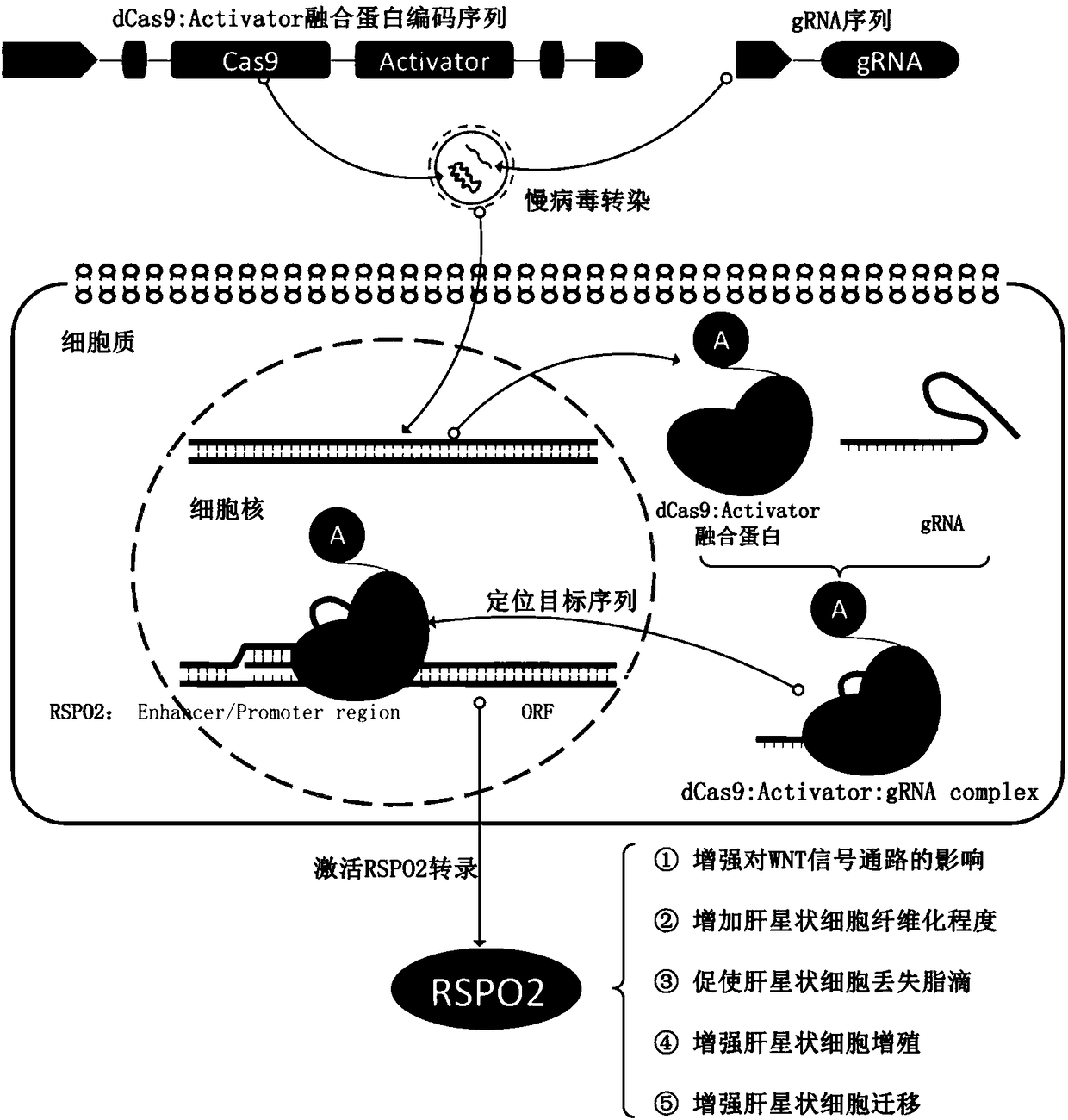
SgRNA specifically targeting human RSPO2 gene in CRISPR-Cas9 system and activation method and application thereof
A specific, genetic technology that can be used in retro-RNA viruses, DNA/RNA fragments, other methods of inserting foreign genetic material, etc., and can solve problems such as adverse biological effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] Example 1 sgRNA sequence design
[0085] There are currently no clear principles for sgRNA design to ensure efficient and specific activation of target gene (CRISPRa) expression. According to our previous work and experience, the design of the sgRNA sequence in the present invention is as follows: (1) the length of the sgRNA is 20 base sequences; (2) the target site of the sgRNA that specifically activates the human RSPO2 gene is located on the RSPO2 gene. (3) choose to design the target at -120~-75 upstream of the RSPO2 gene transcription initiation site; (4) in the RSPO promoter region, select PAM as 5'-NGG; (5) to make the vector The U6 promoter is valid, so ensure that the sgRNA target sequence starts with G; (6) The format of the sgRNA target sequence is:
[0086] 5`-G-(19N)-NGG-3`(sgRNA target sequence starts with G)
[0087] Or 5`-(20N)-NGG-3`(sgRNA target sequence does not start with G)
[0088] In the above target sequence format, 19N or 20N represents the...
Embodiment 2
[0090] Example 2 sgRNA sequence selection
[0091]Use Blast (www.ncbi.nlm.nig.gov / Blast) to perform homology analysis on the candidate sgRNA sequence and the genome database to ensure that the target sequence of the designed sgRNA is unique and will not be homologous to other gene sequences except the human RSPO2 gene . At the same time, the sgRNA was screened according to the following principles to obtain the sgRNA sequence that efficiently and specifically activates the human RSPO2 gene: (1) the sgRNA target is located at the DNase hypersensitive site; (2) the sgRNA target is located at the transcription initiation site of the RSPO2 gene Upstream -120~-75; (3) Low off-target rate
[0092] According to the above method, among the 10 sgRNAs targeting the human RSPO2 gene, only 3 sgRNA sequences (corresponding to SEQ ID NO. , 6), the target sequence of the sgRNA and the corresponding PAM sequence are shown in Table 1 (corresponding to SEQ ID NO.1, 3, 5 in the sequence list...
Embodiment 3
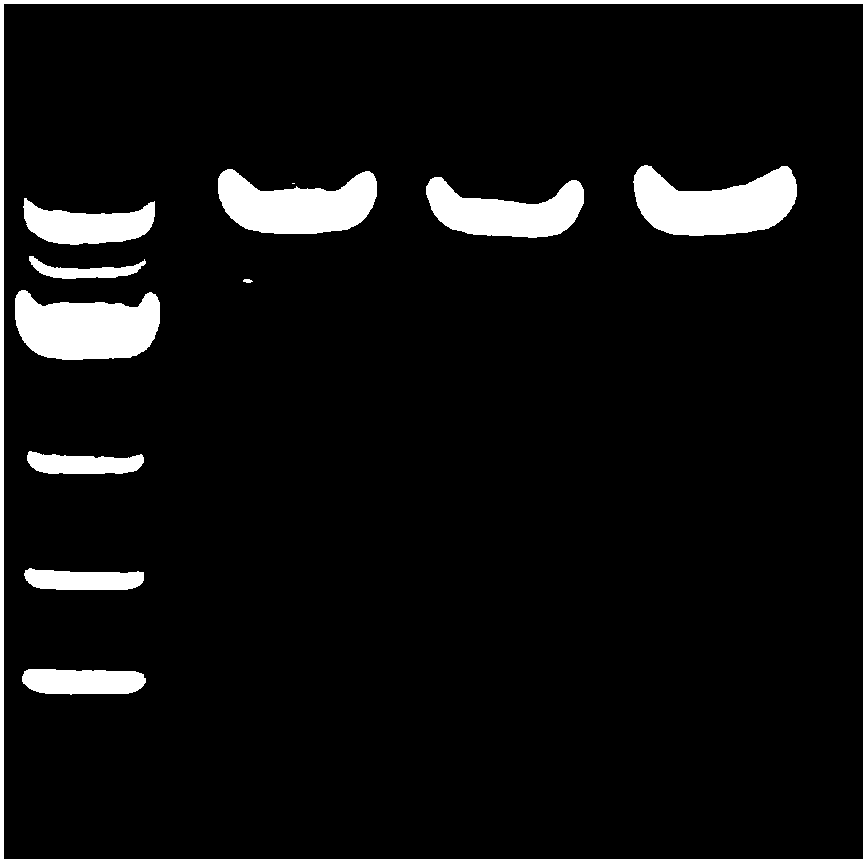
[0093] Example 3 sgRNA oligonucleotide synthesis
[0094] For the above sgRNA sequence targeting human RSPO2 gene, add BsmBI restriction site and PAM sequence at both ends: (1) According to the selected sgRNA, add CACC (BsmBI restriction site sticky Complementary sequence at the end) and G (to ensure that the U6 promoter is effective) to obtain a forward oligonucleotide; (2) According to the selected sgRNA, obtain the complementary strand of DNA corresponding to its target sequence, and add AAAC at its 5' end ( The complementary sequence of the cohesive end of the BsmBI restriction site), add C at the 3' end to get the reverse oligonucleotide
[0095] The above-mentioned forward oligonucleotide and reverse oligonucleotide were respectively synthesized by chemical synthesis method, and the obtained oligonucleotide sequences are shown in Table 2.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com