Brachypodium distachyon drought-resistant gene and expression vector as well as coding protein and application thereof
A technology for expressing vectors and proteins, which is applied in the direction of introducing foreign genetic material, application, and genetic engineering by using vectors. It can solve the problems of crop yield reduction and harm to the normal growth and development of plants, so as to reduce oxidative damage, improve tolerance, and osmotic pressure. Improved effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
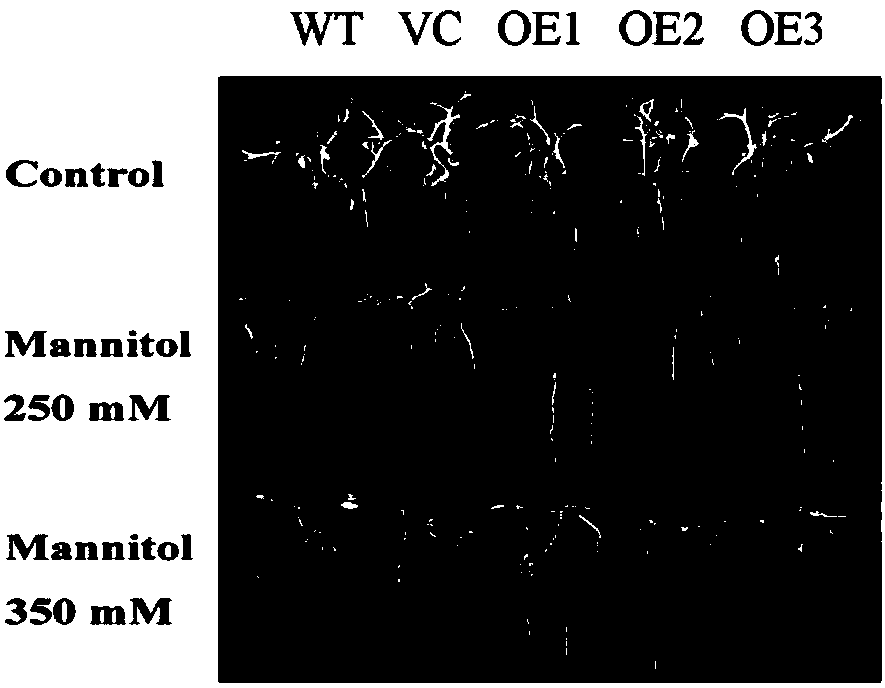
Examples
Embodiment 1
[0033] Example 1: Isolation of the Brachypodium distachyon BdGF14g gene
[0034] 1. Cloning of BdGF14g gene
[0035] The 14-3-3 gene sequence obtained by Phytozome v11.0 (https: / / phytozome.jgi.doe.gov / pz / portal.html) Blast is submitted to NCBI (https: / / www.ncbi.nlm.nih.gov) ) for BLAST comparison analysis. Use the Oligo7 primer design software to specifically amplify the BdGF14g gene of Brachypodium distachyon. The primer sequences are shown in SEQ ID NO.3 and SEQ ID NO.4. The PCR reaction system is shown in Table 1. The PCR program is as follows: pre-denaturation at 98°C for 2 minutes; Denaturation at 98°C for 10s; annealing at 65°C for 5s; extension at 72°C for 11s; final extension at 72°C for 10min; storage at 4°C. Among them, the three consecutive steps of denaturation, annealing and extension set up 35 cyclic reactions.
[0036] Table 1 PCR reaction system
[0037]
[0038] 2. Gel recovery of target fragments
[0039] Spot the PCR reaction product on 1% agarose ge...
Embodiment 2
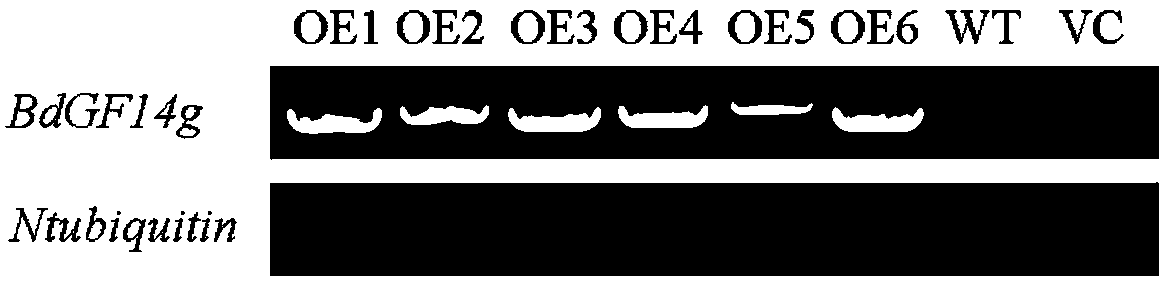
[0062] Example 2: Construction of pBI121-BdGF14g-GFP eukaryotic expression vector
[0063] 1. PCR amplification of BdGF14g gene
[0064] Using the cloning vector pMD18-T-BdGF14g containing the correctly sequenced Brachypodium distachyon BdGF14g gene as a template, use the Oligo7 primer design software to design the XbaI restriction site and BamHI enzyme in the 5' end of the pBI121 vector multiple cloning site region The gene-specific primers at the cleavage site were used for PCR amplification. The primer sequences are shown in Tables SEQ ID NO.5 and SEQ ID NO.6. The PCR reaction system is shown in Table 3 / same as Table 1. The PCR program is as follows: pre-denaturation at 98°C for 2 minutes; 98°C Denaturation at ℃ for 10s; annealing at 65℃ for 5s; extension at 72℃ for 11s; final extension at 72℃ for 10min; storage at 4℃. Among them, the three consecutive steps of denaturation, annealing and extension set up 35 cyclic reactions.
[0065] Table 3 PCR reaction system
[0066]...
Embodiment 3
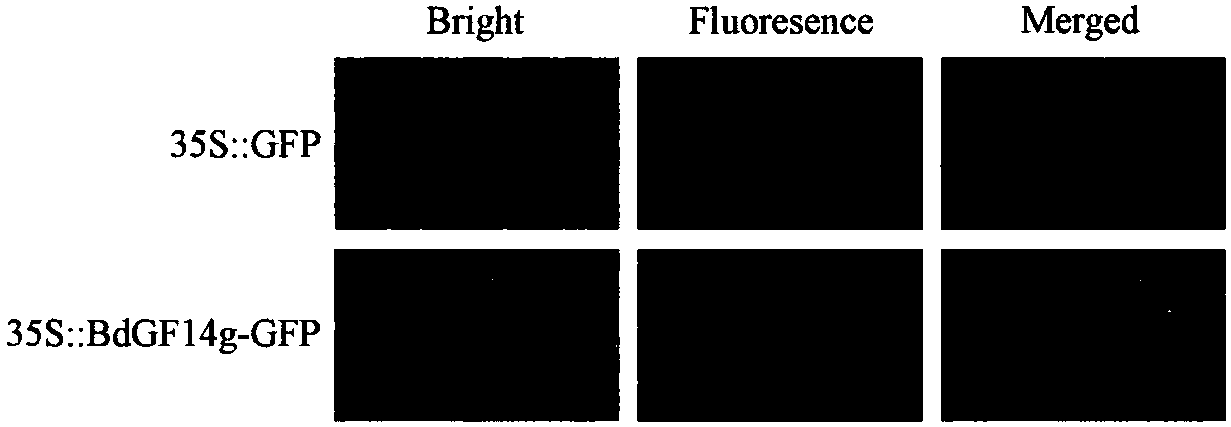
[0113] Example 3: Subcellular localization of Brachypodium distachyon BdGF14g protein
[0114] 1. Preparation and transformation of Agrobacterium competent
[0115] All the operation steps of the preparation and transformation experiment of Agrobacterium competent cells were carried out aseptically in the ultra-clean workbench. The specific process is as follows:
[0116] 1) Use a micropipette to draw 10 μL of the preserved Agrobacterium strain EHA105 from a -80°C ultra-low temperature refrigerator into a 2mL sterilized centrifuge tube, add 1mL of YEB liquid medium containing 50mg / L streptomycin, and store at 28°C Cultivate overnight on a shaker at 200 rpm;
[0117] 2) Take 1 mL of the above-mentioned bacterial solution and transfer it to 100 mL of YEB (containing 50 mg / L streptomycin), expand the culture on a shaker at 28 ° C at 200 rpm, and measure its OD value to OD with a UV / Vis spectrophotometer 600 = about 0.4;
[0118] 3) Ice-bath the above-mentioned Agrobacterium ba...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com