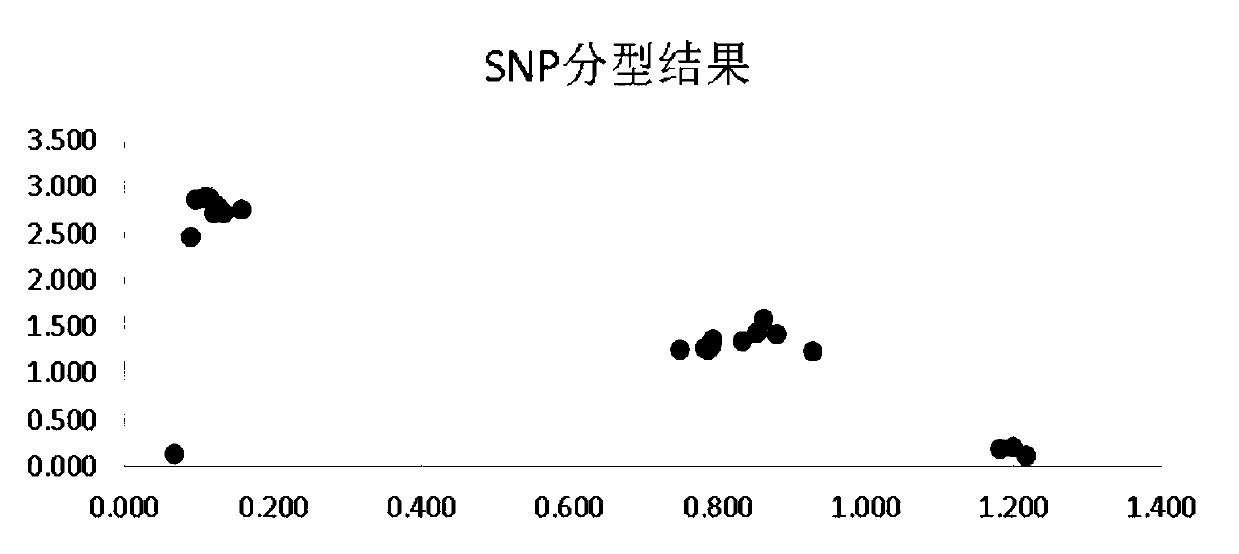
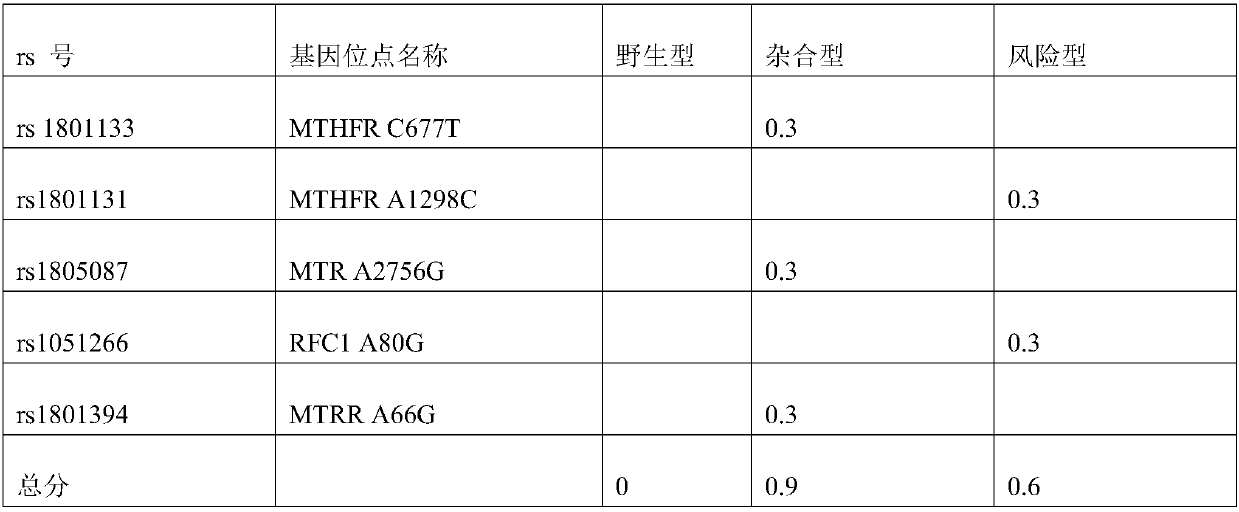
Individual nutrients deficiency risk assessment method based on SNP (Single Nucleotide Polymorphism) site
A risk assessment and risk assessment model technology, applied in the field of human nutrition assessment, can solve problems such as unfavorable automation and long running time, and achieve the effect of ensuring accuracy, strong ductility, and flexible customization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Extraction and quality control of embodiment 1 human DNA
[0050] Use a DNA extraction kit or salting-out method to extract human DNA from blood or other tissues according to the instructions, and use a DNA detector or (and) agarose gel electrophoresis to test the quality of the DNA sample.
Embodiment 2
[0051] Design and synthesis of embodiment 2 SNP site primers
[0052] The first primer mixture contains two specific upstream primers and one universal downstream primer, and the fluorescent substance mixture contains two primers with fluorescent groups. The SNP site in the primer mixture is located at the last base at the 3' end of the upstream primer, and the tag sequences at the 5' end of the two specific upstream primers are different. During PCR amplification, the two specific primers competitively bind. The two primers with fluorescent groups in the fluorescent substance mixture are complementary to the 5' tag sequences of the two specific upstream primers in the primer mixture. Before the PCR reaction, the fluorescent groups (FAM or VIC) do not fluoresce and cannot be detected. Signal, during the PCR process, a sequence that is completely complementary to the upstream primer is synthesized, the fluorescent primer can be combined with the DNA sequence that is completely ...
Embodiment 3
[0080] Design and synthesis of embodiment 3SNP site primers
[0081] 1. Pre-spot primers or DNA on each well of the chip or well plate using a manual or automatic spotting instrument.
[0082] 2. Inject the master mix into the injection hole of the chip or each well of the orifice plate
[0083] The master mix includes 4 μl of DNA or primer mixture with a concentration of 2.5-50 ng / μl, 10 μl of fluorescent substance mixture, and 6 μl of ultrapure water. All components need to be vortexed and mixed evenly before use, and then the reaction master mix is manually added from the chip injection hole or automatic sampler to the chip or well plate. For 96-well plate, 384-well plate and 1536-well plate, the volume of each individual reaction well is 10μl, 5μl and 1μl.
[0084] 3. Seal the membrane and centrifuge
[0085] Blocks were sealed and centrifuged. Each type of orifice plate is sealed and centrifuged manually or by heat sealing machine and (or) laser sealing machine.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com