GRNA, expression vector, knockout system and kit for knocking out CXCR4 gene
A technology of expression vectors and kits, applied in the field of genetic engineering, can solve problems such as high cost of use, cumbersome and complicated design and use, and achieve the effects of improving feasibility, reducing safety risks, and reducing off-target efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 gRNA preliminary screening
[0035] 1. gRNA preparation
[0036] (1) designing a 20nt gRNA sequence according to the sequence of the CXCR4 gene, the target sequence of the gRNA is shown in one of SEQ ID NO:1-SEQ ID NO:24;
[0037] (2) Synthesize the sense strand and antisense strand of the gRNA target sequence respectively (add cacc at the 5'-end of the sense strand, if the first nucleotide at the 5'-end of the sense strand is not guanine G, then add cacc at the 5'-end of the sense strand Add caccG at the -end; add aaac at the 5'-end of the antisense strand, if the first nucleotide at the 5'-end of the sense strand is not guanine G, then add C at the 3'-end of the antisense strand);
[0038] (3) The above-mentioned gRNA sense strand and antisense strand were mixed, treated at 90°C, cooled naturally to room temperature for annealing treatment, and double-stranded gRNA with sticky ends was synthesized.
[0039] 2. Carrier preparation
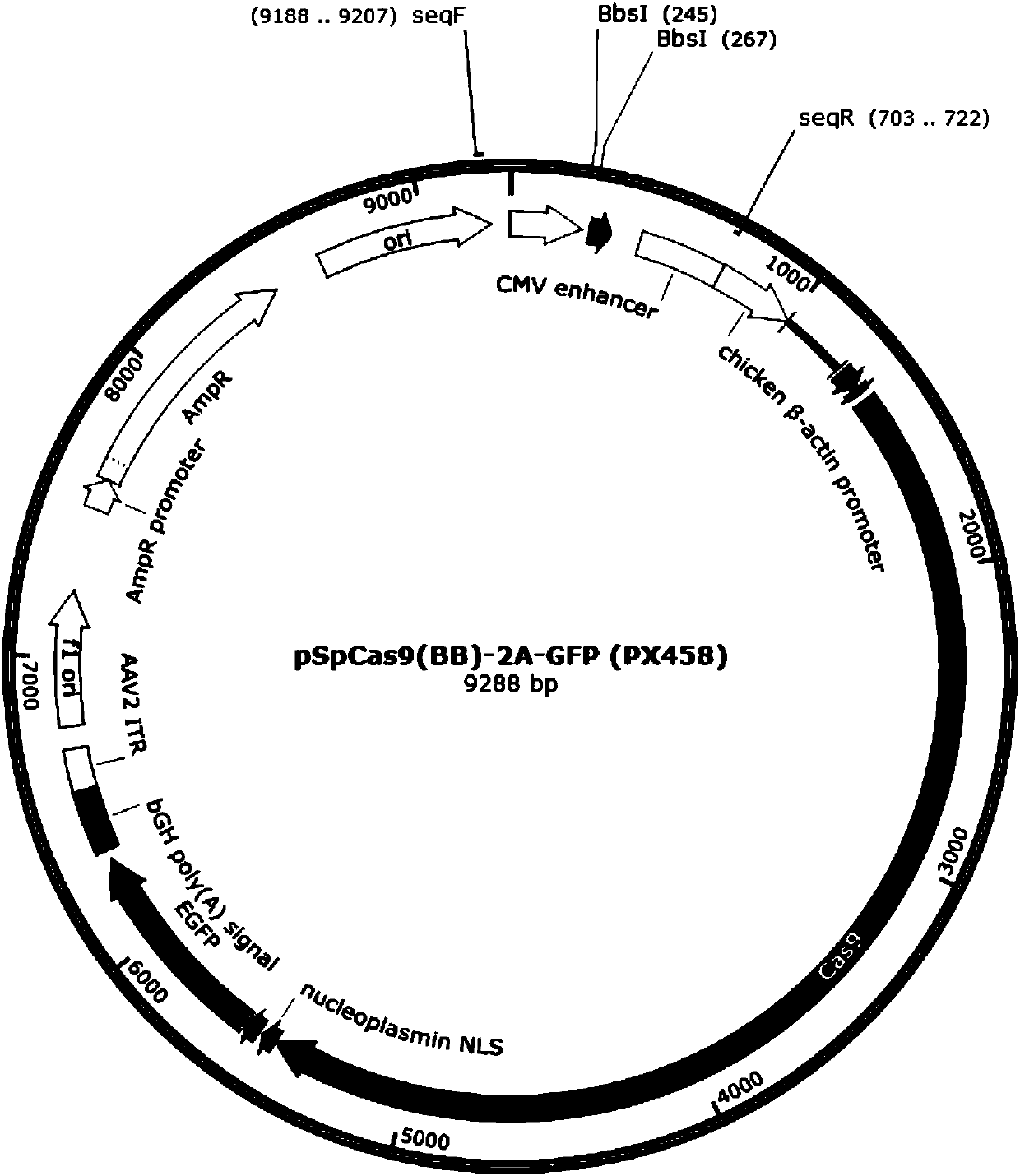
[0040] (1)pX458( figu...
Embodiment 2
[0066] Embodiment 2 gRNA combinatorial screening
[0067] 1. Preparation of gRNA combination
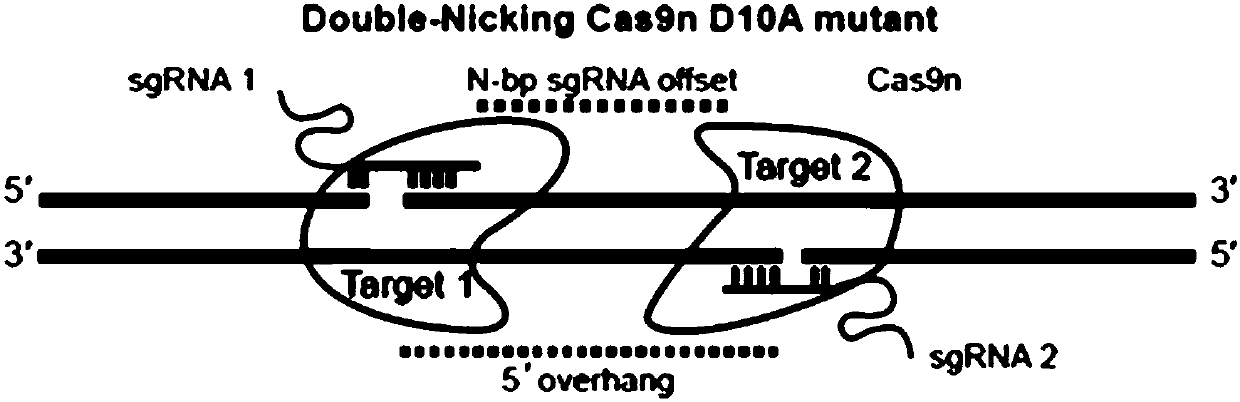
[0068] (1) Select the gRNA with higher mutation efficiency analyzed by T7E1 in Example 1, and select a paired gRNA within 200 bp of the DNA complementary strand of the target sequence it binds to, and the paired combination is shown in Table 2;
[0069] Table 2 gRNA pairing combination
[0070]
[0071]
[0072] (2) From the gRNA sense strand and antisense strand synthesized in 1 (2) of Example 1, select the fragments required for the above pairing, and anneal according to the method of 1 (3) of Example 1 to synthesize the double gRNA with cohesive ends. stranded gRNA.
[0073] 2. Carrier preparation
[0074] (1) pX461 plasmid (such as Figure 4 ) amplify and extract, and measure the plasmid concentration;
[0075] (2) Digest pX461 with restriction endonuclease Bbs I, digest at 37°C for 1 hour, and then add loading buffer to terminate the reaction.
[0076] (3) The linear...
Embodiment 3
[0097] Embodiment 3 and the contrast of prior art
[0098] 1. Preparation of gRNA
[0099] (1) The sense strand and antisense strand of the target sequence corresponding to the gRNA with the highest mutation efficiency in the outsourced synthesis comparison file (add cacc to the 5'-end of the sense strand, if the first nucleotide at the 5'-end of the sense strand is not Guanine G, add caccG at the 5'-end of the sense strand; add aaac at the 5'-end of the antisense strand, if the first nucleotide at the 5'-end of the sense strand is not guanine G, then add aaac at the 5'-end of the antisense strand The 3'-end of the chain adds C), and the DNA sequence of the sense chain is shown in Table 2. In Example 1, gRNAs with higher mutation efficiency analyzed by T7E1 (the corresponding sense strands of the target sequences are SEQ ID NO:7, SEQ ID NO:20, and SEQ ID NO:24) were selected for comparison.
[0100] Table 2 The positive-sense strand of the target sequence corresponding to th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com