Method for preparing arabidopsis autophagy gene mutant and application of arabidopsis autophagy gene mutant
An autophagy gene and mutant technology, applied in the field of preparation of Arabidopsis autophagy gene mutants, can solve problems such as affecting grain quality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
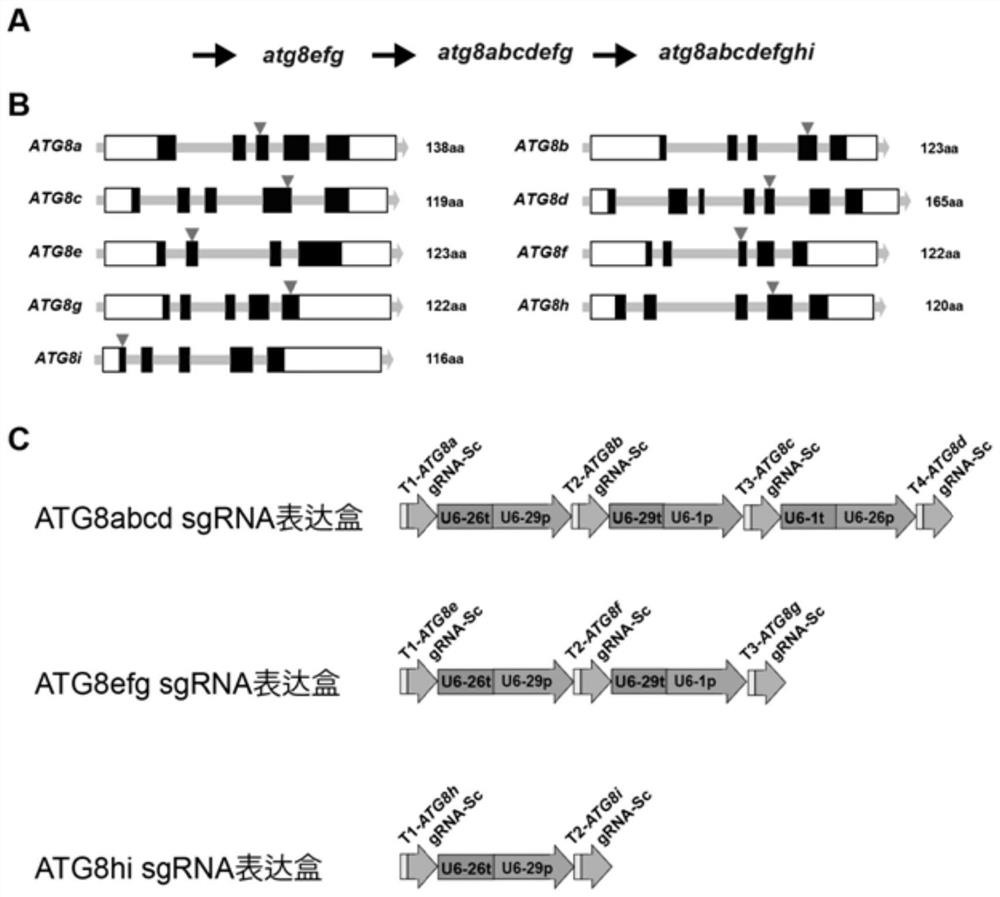
[0040] Embodiment 1 Creation of Arabidopsis nine-fold mutant atg8abcdefghi
[0041] according to figure 1 The technology roadmap shown in A shows that the Arabidopsis atg8abcdefghi nine-mutant homozygous line was constructed using CRISPR-Cas9 gene editing technology. Specific steps are as follows:
[0042] 1. Creation of atg8efg triple mutant
[0043] (1) Obtain the genome sequences of ATG8e (AT2G45170), ATG8f (AT4G16520), and ATG8g (AT3G60640) from the Arabidopsis TAIR database, and use the website http: / / skl.scau.edu.cn / according to the CDS sequence in the genome sequence Determine the CRISPR / Cas9 gene editing site (gene editing site such as figure 1 shown in B).
[0044] The ATG8e gene editing site is located in the third exon, and the sequence is GAAAAGAGAAAAGCTGAAGC (SEQ ID NO.1);
[0045] The ATG8f gene editing site is located in the second exon, and the sequence is GATCCAGGTGATTGTTGAGA (SEQ ID NO.2);
[0046] The ATG8g gene editing site is located in the fifth ex...
Embodiment 2
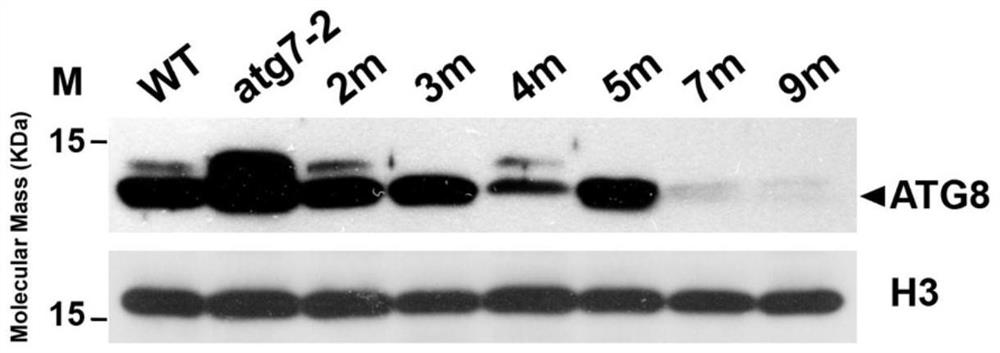
[0137] Example 2 Identification of ATG8 protein expression of Arabidopsis nine-fold mutant atg8abcdefghi by western blot hybridization
[0138] 1. Experimental method
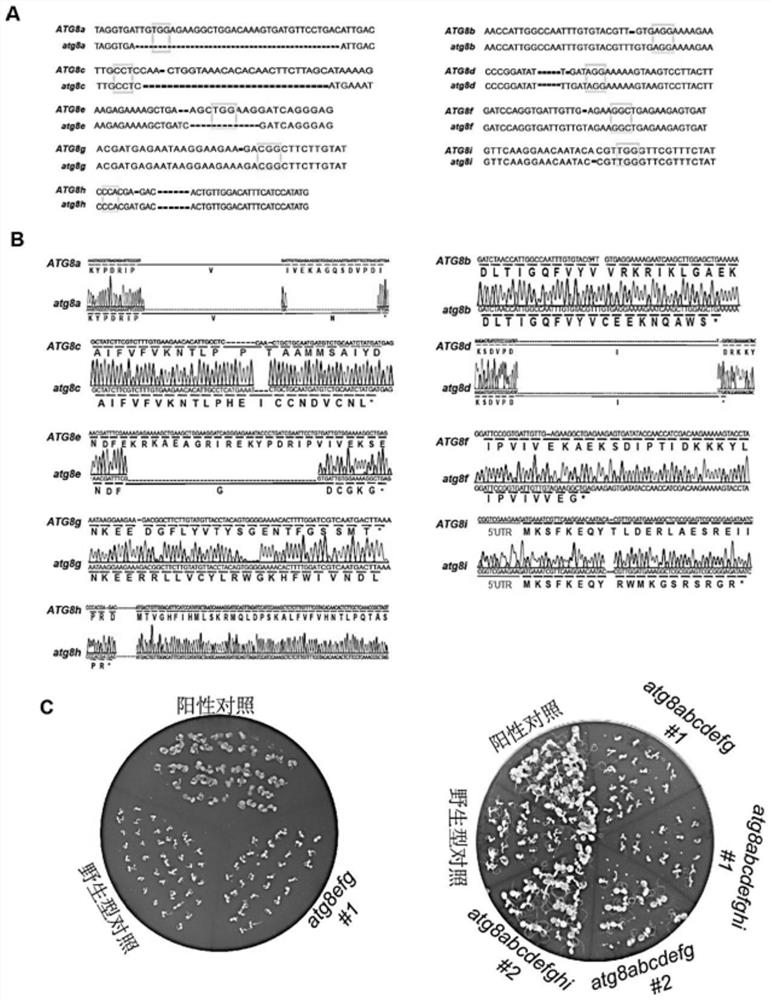
[0139] (1) Protein extraction; wild-type Arabidopsis thaliana, autophagy mutant atg7-2 Arabidopsis (ABRC Arabidopsis Collection Center; article number: GABI_655B06), atg8hi mutant (using wild-type seeds as T0 seeds, using Agrobacterium, obtained by introducing pHEE401E-ATG8hi into wild-type seeds), the atg8efg homozygous triple mutant and atg8abcd mutant prepared in Example 1 (using wild-type seeds as T0 generation seeds, using Agrobacterium to introduce pHEE401E-ATG8abcd into wild type seeds), atg8efghi mutant (atg8efg is T0 generation seed, using Agrobacterium, pHEE401E-ATG8hi is introduced into atg8efg to obtain), atg8abcdefg homozygous seven mutants obtained in Example 1, atg8abcdefghi pure Seeds of Hejiu mutants were grown on Petri dishes for about 1 week. Weigh about 100 mg of the whole Arabidopsis seed...
Embodiment 3
[0144] Example 3 Phenotypic Analysis of Nitrogen Deficiency Stress in Arabidopsis atg8abcdefghi Homozygous Nine Mutants
[0145] 1. Experimental method
[0146] (1) The seeds of wild-type Arabidopsis thaliana, autophagy mutant atg7-2 and atg8abcdefghi homozygous nine mutants prepared in Example 1 were soaked and sterilized with 75% ethanol for 10 min respectively, and the sterilized seeds were sterilized at 4 ℃ refrigerator vernalization for 2 days.
[0147] (2) The vernalized seeds were sown on-demand in 1 / 2 MS solid medium containing 0.7% agarose, and placed in a light incubator at 22°C for germination and culture for 7 days.
[0148] (3) Prepare MS liquid medium (1L): weigh 4.3g MS solid medium (Sigma, M5519-1L), 0.4g of MES (Sangon Bioengineering (Shanghai) Co., Ltd., A100169), 10g of sucrose , with ddH 2 O was adjusted to 1 L, and finally the pH value was adjusted to 5.7 with 2 mM KOH.
[0149] Prepare MS-N liquid medium (1L): Measure 100mL of 10×MS-N solution (Sigma,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com